
Erwinia iniecta
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Erwiniaceae; Erwinia
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
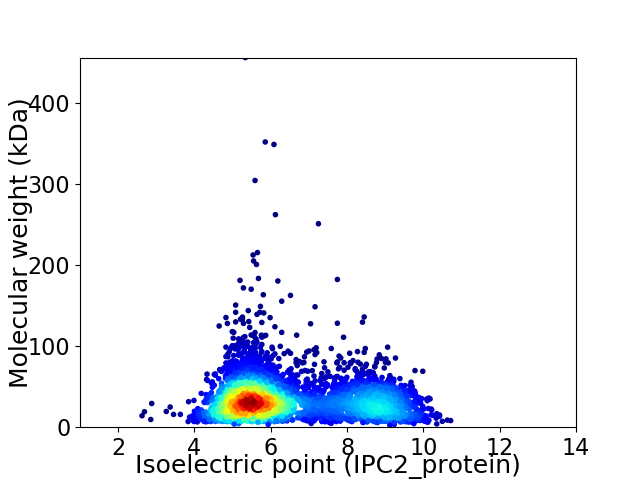
Virtual 2D-PAGE plot for 4105 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0L7THG5|A0A0L7THG5_9GAMM Lipoprotein OS=Erwinia iniecta OX=1560201 GN=NG42_09210 PE=4 SV=1
MM1 pKa = 7.73AYY3 pKa = 10.03EE4 pKa = 4.21EE5 pKa = 4.33LLEE8 pKa = 4.18EE9 pKa = 4.15QRR11 pKa = 11.84EE12 pKa = 4.19EE13 pKa = 3.64TRR15 pKa = 11.84LIIEE19 pKa = 4.54EE20 pKa = 4.2LLEE23 pKa = 5.25DD24 pKa = 4.98GSDD27 pKa = 3.55PDD29 pKa = 3.34ALYY32 pKa = 10.6AIEE35 pKa = 5.05HH36 pKa = 6.49HH37 pKa = 6.6LSCNNFDD44 pKa = 3.91SLEE47 pKa = 4.1KK48 pKa = 10.56VAVDD52 pKa = 3.67AFKK55 pKa = 10.92LGYY58 pKa = 9.35EE59 pKa = 3.98VSEE62 pKa = 4.28PEE64 pKa = 4.5EE65 pKa = 5.14LDD67 pKa = 4.59LEE69 pKa = 5.18DD70 pKa = 3.82GTKK73 pKa = 10.66VMCCDD78 pKa = 4.38IISEE82 pKa = 4.33GALNAEE88 pKa = 4.91LIDD91 pKa = 3.97AQVEE95 pKa = 4.18QLVILAAKK103 pKa = 10.37YY104 pKa = 10.34NVDD107 pKa = 3.26YY108 pKa = 10.96DD109 pKa = 3.31GWGTYY114 pKa = 10.13FEE116 pKa = 6.04DD117 pKa = 5.39PDD119 pKa = 3.93AQDD122 pKa = 4.67DD123 pKa = 3.9EE124 pKa = 5.75GEE126 pKa = 4.36EE127 pKa = 4.27GDD129 pKa = 5.62EE130 pKa = 4.68VDD132 pKa = 4.95EE133 pKa = 5.48DD134 pKa = 4.13DD135 pKa = 4.04TGVRR139 pKa = 11.84HH140 pKa = 6.33
MM1 pKa = 7.73AYY3 pKa = 10.03EE4 pKa = 4.21EE5 pKa = 4.33LLEE8 pKa = 4.18EE9 pKa = 4.15QRR11 pKa = 11.84EE12 pKa = 4.19EE13 pKa = 3.64TRR15 pKa = 11.84LIIEE19 pKa = 4.54EE20 pKa = 4.2LLEE23 pKa = 5.25DD24 pKa = 4.98GSDD27 pKa = 3.55PDD29 pKa = 3.34ALYY32 pKa = 10.6AIEE35 pKa = 5.05HH36 pKa = 6.49HH37 pKa = 6.6LSCNNFDD44 pKa = 3.91SLEE47 pKa = 4.1KK48 pKa = 10.56VAVDD52 pKa = 3.67AFKK55 pKa = 10.92LGYY58 pKa = 9.35EE59 pKa = 3.98VSEE62 pKa = 4.28PEE64 pKa = 4.5EE65 pKa = 5.14LDD67 pKa = 4.59LEE69 pKa = 5.18DD70 pKa = 3.82GTKK73 pKa = 10.66VMCCDD78 pKa = 4.38IISEE82 pKa = 4.33GALNAEE88 pKa = 4.91LIDD91 pKa = 3.97AQVEE95 pKa = 4.18QLVILAAKK103 pKa = 10.37YY104 pKa = 10.34NVDD107 pKa = 3.26YY108 pKa = 10.96DD109 pKa = 3.31GWGTYY114 pKa = 10.13FEE116 pKa = 6.04DD117 pKa = 5.39PDD119 pKa = 3.93AQDD122 pKa = 4.67DD123 pKa = 3.9EE124 pKa = 5.75GEE126 pKa = 4.36EE127 pKa = 4.27GDD129 pKa = 5.62EE130 pKa = 4.68VDD132 pKa = 4.95EE133 pKa = 5.48DD134 pKa = 4.13DD135 pKa = 4.04TGVRR139 pKa = 11.84HH140 pKa = 6.33
Molecular weight: 15.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0L7T779|A0A0L7T779_9GAMM Uncharacterized protein OS=Erwinia iniecta OX=1560201 GN=NG42_05255 PE=4 SV=1
MM1 pKa = 7.54SLPPSANIFTVSRR14 pKa = 11.84LNTTVRR20 pKa = 11.84KK21 pKa = 9.2LLEE24 pKa = 4.02MEE26 pKa = 4.85MGQIWLSAEE35 pKa = 3.63ISNFSQPSSGHH46 pKa = 5.4WYY48 pKa = 7.9FTLKK52 pKa = 10.77DD53 pKa = 3.34DD54 pKa = 4.55GAQVRR59 pKa = 11.84CAMFRR64 pKa = 11.84NSNRR68 pKa = 11.84RR69 pKa = 11.84VTFRR73 pKa = 11.84PQNGQQVLVRR83 pKa = 11.84ASITLYY89 pKa = 10.38EE90 pKa = 4.03PRR92 pKa = 11.84GDD94 pKa = 3.63YY95 pKa = 10.67QLIAEE100 pKa = 4.52SMQPAGDD107 pKa = 4.14GLLQQQFEE115 pKa = 4.25QLKK118 pKa = 9.82QRR120 pKa = 11.84LAAEE124 pKa = 4.42GLFDD128 pKa = 4.35QQFKK132 pKa = 10.98QPLPDD137 pKa = 3.79PARR140 pKa = 11.84QVGVITSATGAALHH154 pKa = 6.59DD155 pKa = 4.26VLRR158 pKa = 11.84VLHH161 pKa = 6.73RR162 pKa = 11.84RR163 pKa = 11.84DD164 pKa = 3.07PSLPVIIYY172 pKa = 6.4PTQVQGADD180 pKa = 3.3APASLVRR187 pKa = 11.84AIEE190 pKa = 4.09LANQRR195 pKa = 11.84GEE197 pKa = 4.18CDD199 pKa = 3.34VLIVGRR205 pKa = 11.84GGGSLEE211 pKa = 4.7DD212 pKa = 2.99LWSFNDD218 pKa = 3.26EE219 pKa = 3.92RR220 pKa = 11.84VARR223 pKa = 11.84AIFASRR229 pKa = 11.84IPVVSAVGHH238 pKa = 5.22EE239 pKa = 4.23TDD241 pKa = 3.12VTIADD246 pKa = 4.4FVADD250 pKa = 4.65LRR252 pKa = 11.84APTPSAAAEE261 pKa = 3.91LVSRR265 pKa = 11.84NQIEE269 pKa = 4.53RR270 pKa = 11.84LRR272 pKa = 11.84QLQSQQQRR280 pKa = 11.84MEE282 pKa = 4.04MALDD286 pKa = 3.53YY287 pKa = 11.32YY288 pKa = 10.83LAQKK292 pKa = 9.87QRR294 pKa = 11.84AFTQLHH300 pKa = 6.35HH301 pKa = 7.37RR302 pKa = 11.84LQQQHH307 pKa = 5.78PQLRR311 pKa = 11.84LARR314 pKa = 11.84QQTSLFRR321 pKa = 11.84LQRR324 pKa = 11.84RR325 pKa = 11.84LDD327 pKa = 3.53DD328 pKa = 4.16AMQSRR333 pKa = 11.84LRR335 pKa = 11.84QSLRR339 pKa = 11.84QQEE342 pKa = 4.51RR343 pKa = 11.84IEE345 pKa = 3.98QRR347 pKa = 11.84LAAQQPQGRR356 pKa = 11.84LHH358 pKa = 7.48RR359 pKa = 11.84AQQQLQQWQYY369 pKa = 11.16RR370 pKa = 11.84LQQGMQSQLSRR381 pKa = 11.84DD382 pKa = 3.44KK383 pKa = 11.18QRR385 pKa = 11.84FGTLAAQLEE394 pKa = 5.05GVSPLATLARR404 pKa = 11.84GFSVTTAGDD413 pKa = 3.73GQVVRR418 pKa = 11.84KK419 pKa = 7.74TRR421 pKa = 11.84QIKK424 pKa = 10.62RR425 pKa = 11.84GDD427 pKa = 3.65TLKK430 pKa = 10.02TRR432 pKa = 11.84LDD434 pKa = 4.22DD435 pKa = 3.93GWVEE439 pKa = 4.22SQVTRR444 pKa = 11.84VSAIKK449 pKa = 10.52KK450 pKa = 6.81KK451 pKa = 8.64TRR453 pKa = 3.14
MM1 pKa = 7.54SLPPSANIFTVSRR14 pKa = 11.84LNTTVRR20 pKa = 11.84KK21 pKa = 9.2LLEE24 pKa = 4.02MEE26 pKa = 4.85MGQIWLSAEE35 pKa = 3.63ISNFSQPSSGHH46 pKa = 5.4WYY48 pKa = 7.9FTLKK52 pKa = 10.77DD53 pKa = 3.34DD54 pKa = 4.55GAQVRR59 pKa = 11.84CAMFRR64 pKa = 11.84NSNRR68 pKa = 11.84RR69 pKa = 11.84VTFRR73 pKa = 11.84PQNGQQVLVRR83 pKa = 11.84ASITLYY89 pKa = 10.38EE90 pKa = 4.03PRR92 pKa = 11.84GDD94 pKa = 3.63YY95 pKa = 10.67QLIAEE100 pKa = 4.52SMQPAGDD107 pKa = 4.14GLLQQQFEE115 pKa = 4.25QLKK118 pKa = 9.82QRR120 pKa = 11.84LAAEE124 pKa = 4.42GLFDD128 pKa = 4.35QQFKK132 pKa = 10.98QPLPDD137 pKa = 3.79PARR140 pKa = 11.84QVGVITSATGAALHH154 pKa = 6.59DD155 pKa = 4.26VLRR158 pKa = 11.84VLHH161 pKa = 6.73RR162 pKa = 11.84RR163 pKa = 11.84DD164 pKa = 3.07PSLPVIIYY172 pKa = 6.4PTQVQGADD180 pKa = 3.3APASLVRR187 pKa = 11.84AIEE190 pKa = 4.09LANQRR195 pKa = 11.84GEE197 pKa = 4.18CDD199 pKa = 3.34VLIVGRR205 pKa = 11.84GGGSLEE211 pKa = 4.7DD212 pKa = 2.99LWSFNDD218 pKa = 3.26EE219 pKa = 3.92RR220 pKa = 11.84VARR223 pKa = 11.84AIFASRR229 pKa = 11.84IPVVSAVGHH238 pKa = 5.22EE239 pKa = 4.23TDD241 pKa = 3.12VTIADD246 pKa = 4.4FVADD250 pKa = 4.65LRR252 pKa = 11.84APTPSAAAEE261 pKa = 3.91LVSRR265 pKa = 11.84NQIEE269 pKa = 4.53RR270 pKa = 11.84LRR272 pKa = 11.84QLQSQQQRR280 pKa = 11.84MEE282 pKa = 4.04MALDD286 pKa = 3.53YY287 pKa = 11.32YY288 pKa = 10.83LAQKK292 pKa = 9.87QRR294 pKa = 11.84AFTQLHH300 pKa = 6.35HH301 pKa = 7.37RR302 pKa = 11.84LQQQHH307 pKa = 5.78PQLRR311 pKa = 11.84LARR314 pKa = 11.84QQTSLFRR321 pKa = 11.84LQRR324 pKa = 11.84RR325 pKa = 11.84LDD327 pKa = 3.53DD328 pKa = 4.16AMQSRR333 pKa = 11.84LRR335 pKa = 11.84QSLRR339 pKa = 11.84QQEE342 pKa = 4.51RR343 pKa = 11.84IEE345 pKa = 3.98QRR347 pKa = 11.84LAAQQPQGRR356 pKa = 11.84LHH358 pKa = 7.48RR359 pKa = 11.84AQQQLQQWQYY369 pKa = 11.16RR370 pKa = 11.84LQQGMQSQLSRR381 pKa = 11.84DD382 pKa = 3.44KK383 pKa = 11.18QRR385 pKa = 11.84FGTLAAQLEE394 pKa = 5.05GVSPLATLARR404 pKa = 11.84GFSVTTAGDD413 pKa = 3.73GQVVRR418 pKa = 11.84KK419 pKa = 7.74TRR421 pKa = 11.84QIKK424 pKa = 10.62RR425 pKa = 11.84GDD427 pKa = 3.65TLKK430 pKa = 10.02TRR432 pKa = 11.84LDD434 pKa = 4.22DD435 pKa = 3.93GWVEE439 pKa = 4.22SQVTRR444 pKa = 11.84VSAIKK449 pKa = 10.52KK450 pKa = 6.81KK451 pKa = 8.64TRR453 pKa = 3.14
Molecular weight: 51.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1321614 |
23 |
4133 |
322.0 |
35.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.892 ± 0.044 | 0.983 ± 0.013 |
5.222 ± 0.029 | 5.447 ± 0.035 |
3.876 ± 0.027 | 7.266 ± 0.038 |
2.246 ± 0.018 | 5.769 ± 0.034 |
4.032 ± 0.032 | 11.173 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.707 ± 0.018 | 3.739 ± 0.028 |
4.459 ± 0.025 | 5.024 ± 0.039 |
5.707 ± 0.029 | 6.255 ± 0.031 |
5.096 ± 0.024 | 6.846 ± 0.031 |
1.515 ± 0.016 | 2.737 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
