
Morelia spilota papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyomupapillomavirus; Dyomupapillomavirus 1
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
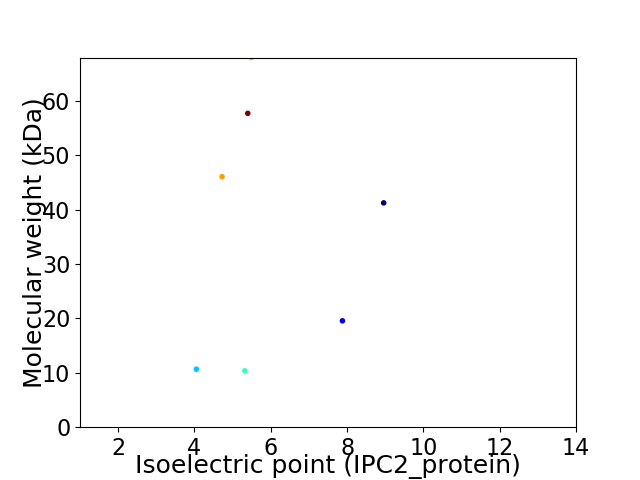
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|G3DRD2|G3DRD2_9PAPI Replication protein E1 OS=Morelia spilota papillomavirus 1 OX=1081054 GN=E1 PE=3 SV=1
MM1 pKa = 7.5MIGPKK6 pKa = 10.06LDD8 pKa = 3.44VDD10 pKa = 4.26LLCYY14 pKa = 10.43EE15 pKa = 5.39DD16 pKa = 4.24LTSPNPDD23 pKa = 3.05EE24 pKa = 4.71AAIPAPPPSPSHH36 pKa = 6.73PDD38 pKa = 2.99LRR40 pKa = 11.84AYY42 pKa = 9.56TIDD45 pKa = 4.61LGCGYY50 pKa = 8.1CTKK53 pKa = 10.22PLRR56 pKa = 11.84FVTVATAEE64 pKa = 4.49CISLFNRR71 pKa = 11.84LLLLDD76 pKa = 5.39LYY78 pKa = 9.98ILCPEE83 pKa = 4.57CVDD86 pKa = 4.81EE87 pKa = 6.41RR88 pKa = 11.84DD89 pKa = 2.93LDD91 pKa = 3.93YY92 pKa = 11.46YY93 pKa = 11.49YY94 pKa = 11.33GGG96 pKa = 3.81
MM1 pKa = 7.5MIGPKK6 pKa = 10.06LDD8 pKa = 3.44VDD10 pKa = 4.26LLCYY14 pKa = 10.43EE15 pKa = 5.39DD16 pKa = 4.24LTSPNPDD23 pKa = 3.05EE24 pKa = 4.71AAIPAPPPSPSHH36 pKa = 6.73PDD38 pKa = 2.99LRR40 pKa = 11.84AYY42 pKa = 9.56TIDD45 pKa = 4.61LGCGYY50 pKa = 8.1CTKK53 pKa = 10.22PLRR56 pKa = 11.84FVTVATAEE64 pKa = 4.49CISLFNRR71 pKa = 11.84LLLLDD76 pKa = 5.39LYY78 pKa = 9.98ILCPEE83 pKa = 4.57CVDD86 pKa = 4.81EE87 pKa = 6.41RR88 pKa = 11.84DD89 pKa = 2.93LDD91 pKa = 3.93YY92 pKa = 11.46YY93 pKa = 11.49YY94 pKa = 11.33GGG96 pKa = 3.81
Molecular weight: 10.67 kDa
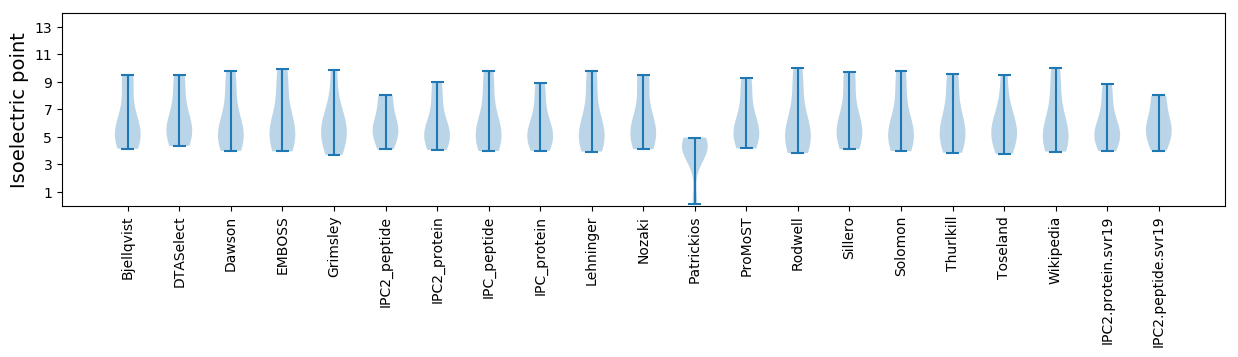
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G3DRD4|G3DRD4_9PAPI E4 (Fragment) OS=Morelia spilota papillomavirus 1 OX=1081054 PE=4 SV=1
MM1 pKa = 6.55EE2 pKa = 4.8TLRR5 pKa = 11.84KK6 pKa = 9.77RR7 pKa = 11.84LDD9 pKa = 3.48AVQEE13 pKa = 4.0ALLDD17 pKa = 3.66IYY19 pKa = 9.82EE20 pKa = 4.37TAEE23 pKa = 4.04NTLEE27 pKa = 4.07TQIKK31 pKa = 8.02HH32 pKa = 4.81WKK34 pKa = 9.55LVRR37 pKa = 11.84QEE39 pKa = 3.65QTLLYY44 pKa = 9.86FARR47 pKa = 11.84QQGLSHH53 pKa = 7.09IGLQTVPALQISEE66 pKa = 4.46SKK68 pKa = 10.63AKK70 pKa = 9.88EE71 pKa = 3.92AIEE74 pKa = 3.76IQLYY78 pKa = 10.58LEE80 pKa = 4.43SLQNSKK86 pKa = 11.17YY87 pKa = 10.59GDD89 pKa = 4.26EE90 pKa = 4.19PWTMQEE96 pKa = 3.75TSALTFFAVPSRR108 pKa = 11.84TFKK111 pKa = 10.72KK112 pKa = 10.38GPTTVVVQFEE122 pKa = 4.33QHH124 pKa = 4.59TQSYY128 pKa = 6.6TTWTYY133 pKa = 11.42LYY135 pKa = 9.61IQTDD139 pKa = 3.07TDD141 pKa = 2.87MWTKK145 pKa = 10.75YY146 pKa = 9.76VGKK149 pKa = 10.31VSNEE153 pKa = 3.29GAYY156 pKa = 10.24YY157 pKa = 10.46SAGGGLKK164 pKa = 9.17TFYY167 pKa = 10.48ISFSAEE173 pKa = 3.39AKK175 pKa = 10.27KK176 pKa = 11.11YY177 pKa = 8.72NTDD180 pKa = 2.72NWTLVYY186 pKa = 10.23KK187 pKa = 9.34QLPVLTSSITPLQSPTGTTDD207 pKa = 3.03TAEE210 pKa = 4.09TQPAGGRR217 pKa = 11.84PKK219 pKa = 10.08RR220 pKa = 11.84RR221 pKa = 11.84YY222 pKa = 9.09GRR224 pKa = 11.84RR225 pKa = 11.84SEE227 pKa = 4.11SPRR230 pKa = 11.84PRR232 pKa = 11.84RR233 pKa = 11.84RR234 pKa = 11.84RR235 pKa = 11.84EE236 pKa = 3.84DD237 pKa = 3.35QEE239 pKa = 4.05ASSPGGVPVAPEE251 pKa = 3.99DD252 pKa = 3.42VGSRR256 pKa = 11.84NRR258 pKa = 11.84TVEE261 pKa = 3.89AKK263 pKa = 10.21RR264 pKa = 11.84SSRR267 pKa = 11.84LARR270 pKa = 11.84LLDD273 pKa = 3.74EE274 pKa = 5.62ARR276 pKa = 11.84DD277 pKa = 3.79PPIVVLKK284 pKa = 10.59GNPNSLKK291 pKa = 10.05CLRR294 pKa = 11.84YY295 pKa = 9.43RR296 pKa = 11.84LRR298 pKa = 11.84HH299 pKa = 5.09QYY301 pKa = 10.66SKK303 pKa = 10.67EE304 pKa = 3.8FQHH307 pKa = 7.38ISTTFQWVDD316 pKa = 3.36TVDD319 pKa = 3.35SARR322 pKa = 11.84LGRR325 pKa = 11.84GRR327 pKa = 11.84MLVMFTDD334 pKa = 4.59DD335 pKa = 3.39AQRR338 pKa = 11.84MHH340 pKa = 6.86FLKK343 pKa = 10.56HH344 pKa = 5.26VPLPKK349 pKa = 10.26HH350 pKa = 4.32ITAFKK355 pKa = 10.08GQLDD359 pKa = 4.86GII361 pKa = 4.5
MM1 pKa = 6.55EE2 pKa = 4.8TLRR5 pKa = 11.84KK6 pKa = 9.77RR7 pKa = 11.84LDD9 pKa = 3.48AVQEE13 pKa = 4.0ALLDD17 pKa = 3.66IYY19 pKa = 9.82EE20 pKa = 4.37TAEE23 pKa = 4.04NTLEE27 pKa = 4.07TQIKK31 pKa = 8.02HH32 pKa = 4.81WKK34 pKa = 9.55LVRR37 pKa = 11.84QEE39 pKa = 3.65QTLLYY44 pKa = 9.86FARR47 pKa = 11.84QQGLSHH53 pKa = 7.09IGLQTVPALQISEE66 pKa = 4.46SKK68 pKa = 10.63AKK70 pKa = 9.88EE71 pKa = 3.92AIEE74 pKa = 3.76IQLYY78 pKa = 10.58LEE80 pKa = 4.43SLQNSKK86 pKa = 11.17YY87 pKa = 10.59GDD89 pKa = 4.26EE90 pKa = 4.19PWTMQEE96 pKa = 3.75TSALTFFAVPSRR108 pKa = 11.84TFKK111 pKa = 10.72KK112 pKa = 10.38GPTTVVVQFEE122 pKa = 4.33QHH124 pKa = 4.59TQSYY128 pKa = 6.6TTWTYY133 pKa = 11.42LYY135 pKa = 9.61IQTDD139 pKa = 3.07TDD141 pKa = 2.87MWTKK145 pKa = 10.75YY146 pKa = 9.76VGKK149 pKa = 10.31VSNEE153 pKa = 3.29GAYY156 pKa = 10.24YY157 pKa = 10.46SAGGGLKK164 pKa = 9.17TFYY167 pKa = 10.48ISFSAEE173 pKa = 3.39AKK175 pKa = 10.27KK176 pKa = 11.11YY177 pKa = 8.72NTDD180 pKa = 2.72NWTLVYY186 pKa = 10.23KK187 pKa = 9.34QLPVLTSSITPLQSPTGTTDD207 pKa = 3.03TAEE210 pKa = 4.09TQPAGGRR217 pKa = 11.84PKK219 pKa = 10.08RR220 pKa = 11.84RR221 pKa = 11.84YY222 pKa = 9.09GRR224 pKa = 11.84RR225 pKa = 11.84SEE227 pKa = 4.11SPRR230 pKa = 11.84PRR232 pKa = 11.84RR233 pKa = 11.84RR234 pKa = 11.84RR235 pKa = 11.84EE236 pKa = 3.84DD237 pKa = 3.35QEE239 pKa = 4.05ASSPGGVPVAPEE251 pKa = 3.99DD252 pKa = 3.42VGSRR256 pKa = 11.84NRR258 pKa = 11.84TVEE261 pKa = 3.89AKK263 pKa = 10.21RR264 pKa = 11.84SSRR267 pKa = 11.84LARR270 pKa = 11.84LLDD273 pKa = 3.74EE274 pKa = 5.62ARR276 pKa = 11.84DD277 pKa = 3.79PPIVVLKK284 pKa = 10.59GNPNSLKK291 pKa = 10.05CLRR294 pKa = 11.84YY295 pKa = 9.43RR296 pKa = 11.84LRR298 pKa = 11.84HH299 pKa = 5.09QYY301 pKa = 10.66SKK303 pKa = 10.67EE304 pKa = 3.8FQHH307 pKa = 7.38ISTTFQWVDD316 pKa = 3.36TVDD319 pKa = 3.35SARR322 pKa = 11.84LGRR325 pKa = 11.84GRR327 pKa = 11.84MLVMFTDD334 pKa = 4.59DD335 pKa = 3.39AQRR338 pKa = 11.84MHH340 pKa = 6.86FLKK343 pKa = 10.56HH344 pKa = 5.26VPLPKK349 pKa = 10.26HH350 pKa = 4.32ITAFKK355 pKa = 10.08GQLDD359 pKa = 4.86GII361 pKa = 4.5
Molecular weight: 41.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2240 |
88 |
596 |
320.0 |
36.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.295 ± 0.447 | 1.741 ± 0.701 |
6.161 ± 0.702 | 6.652 ± 0.723 |
5.045 ± 0.534 | 5.313 ± 0.553 |
1.786 ± 0.313 | 5.536 ± 0.53 |
5.134 ± 1.044 | 9.643 ± 1.205 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.696 ± 0.181 | 4.063 ± 0.465 |
5.804 ± 0.831 | 4.42 ± 0.705 |
6.027 ± 0.717 | 7.187 ± 0.629 |
6.875 ± 0.679 | 5.982 ± 0.714 |
1.205 ± 0.351 | 3.438 ± 0.479 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
