
Natrarchaeobaculum aegyptiacum
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Natrarchaeobaculum
Average proteome isoelectric point is 4.76
Get precalculated fractions of proteins
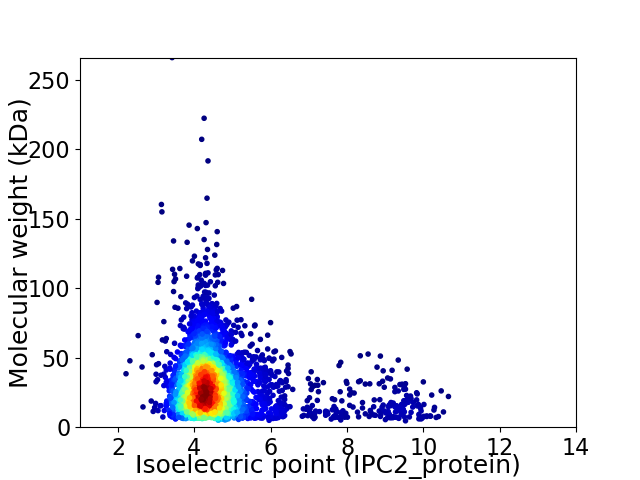
Virtual 2D-PAGE plot for 3626 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z2HVW8|A0A2Z2HVW8_9EURY DUF357 domain-containing protein OS=Natrarchaeobaculum aegyptiacum OX=745377 GN=B1756_17640 PE=4 SV=1
MM1 pKa = 7.44EE2 pKa = 4.86LTWHH6 pKa = 6.23GHH8 pKa = 4.48STWHH12 pKa = 4.8VTVGGTDD19 pKa = 3.66LLIDD23 pKa = 4.15PFFDD27 pKa = 3.84NPKK30 pKa = 9.57TDD32 pKa = 4.65LEE34 pKa = 4.43PADD37 pKa = 4.65VDD39 pKa = 3.95DD40 pKa = 5.39PDD42 pKa = 4.47YY43 pKa = 11.81VLLTHH48 pKa = 6.39GHH50 pKa = 7.0ADD52 pKa = 4.13HH53 pKa = 7.06IAHH56 pKa = 6.9AGEE59 pKa = 4.54FSDD62 pKa = 3.89ATLVATPEE70 pKa = 4.08LVSYY74 pKa = 10.0CQEE77 pKa = 3.68EE78 pKa = 4.66FGFEE82 pKa = 4.05DD83 pKa = 3.69AVGGMGMNLGGTVDD97 pKa = 4.39CGDD100 pKa = 3.57ATVTMVRR107 pKa = 11.84ADD109 pKa = 3.57HH110 pKa = 6.68TNGIMTEE117 pKa = 3.94NDD119 pKa = 3.09ASGGMPAGFVIADD132 pKa = 4.39GDD134 pKa = 4.03PTDD137 pKa = 5.23DD138 pKa = 6.1DD139 pKa = 3.94STTVYY144 pKa = 10.85NAGDD148 pKa = 3.65TALMSEE154 pKa = 4.47MKK156 pKa = 10.54DD157 pKa = 3.43VIGDD161 pKa = 3.88HH162 pKa = 6.89LAPDD166 pKa = 3.87AAIVPIGDD174 pKa = 4.27HH175 pKa = 5.57FTMGPTQAGVAVDD188 pKa = 3.37WTGADD193 pKa = 3.06HH194 pKa = 7.75AFPQHH199 pKa = 6.83YY200 pKa = 8.02DD201 pKa = 3.22TFPPIEE207 pKa = 4.18QDD209 pKa = 3.34PADD212 pKa = 4.07FEE214 pKa = 4.95AAVSDD219 pKa = 4.27AEE221 pKa = 4.43VHH223 pKa = 5.87VLEE226 pKa = 5.26ADD228 pKa = 3.67EE229 pKa = 4.64PFEE232 pKa = 4.37LL233 pKa = 5.69
MM1 pKa = 7.44EE2 pKa = 4.86LTWHH6 pKa = 6.23GHH8 pKa = 4.48STWHH12 pKa = 4.8VTVGGTDD19 pKa = 3.66LLIDD23 pKa = 4.15PFFDD27 pKa = 3.84NPKK30 pKa = 9.57TDD32 pKa = 4.65LEE34 pKa = 4.43PADD37 pKa = 4.65VDD39 pKa = 3.95DD40 pKa = 5.39PDD42 pKa = 4.47YY43 pKa = 11.81VLLTHH48 pKa = 6.39GHH50 pKa = 7.0ADD52 pKa = 4.13HH53 pKa = 7.06IAHH56 pKa = 6.9AGEE59 pKa = 4.54FSDD62 pKa = 3.89ATLVATPEE70 pKa = 4.08LVSYY74 pKa = 10.0CQEE77 pKa = 3.68EE78 pKa = 4.66FGFEE82 pKa = 4.05DD83 pKa = 3.69AVGGMGMNLGGTVDD97 pKa = 4.39CGDD100 pKa = 3.57ATVTMVRR107 pKa = 11.84ADD109 pKa = 3.57HH110 pKa = 6.68TNGIMTEE117 pKa = 3.94NDD119 pKa = 3.09ASGGMPAGFVIADD132 pKa = 4.39GDD134 pKa = 4.03PTDD137 pKa = 5.23DD138 pKa = 6.1DD139 pKa = 3.94STTVYY144 pKa = 10.85NAGDD148 pKa = 3.65TALMSEE154 pKa = 4.47MKK156 pKa = 10.54DD157 pKa = 3.43VIGDD161 pKa = 3.88HH162 pKa = 6.89LAPDD166 pKa = 3.87AAIVPIGDD174 pKa = 4.27HH175 pKa = 5.57FTMGPTQAGVAVDD188 pKa = 3.37WTGADD193 pKa = 3.06HH194 pKa = 7.75AFPQHH199 pKa = 6.83YY200 pKa = 8.02DD201 pKa = 3.22TFPPIEE207 pKa = 4.18QDD209 pKa = 3.34PADD212 pKa = 4.07FEE214 pKa = 4.95AAVSDD219 pKa = 4.27AEE221 pKa = 4.43VHH223 pKa = 5.87VLEE226 pKa = 5.26ADD228 pKa = 3.67EE229 pKa = 4.64PFEE232 pKa = 4.37LL233 pKa = 5.69
Molecular weight: 24.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z2I1A7|A0A2Z2I1A7_9EURY Uncharacterized protein OS=Natrarchaeobaculum aegyptiacum OX=745377 GN=B1756_18615 PE=4 SV=1
MM1 pKa = 7.92RR2 pKa = 11.84FGSDD6 pKa = 2.96RR7 pKa = 11.84SPSVAAVADD16 pKa = 4.12GARR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84PRR23 pKa = 11.84PRR25 pKa = 11.84ATDD28 pKa = 3.37EE29 pKa = 4.6GNGDD33 pKa = 3.54PVRR36 pKa = 11.84RR37 pKa = 11.84TGVCSSDD44 pKa = 2.98GVTPQKK50 pKa = 10.2WPPQRR55 pKa = 11.84NSKK58 pKa = 9.87LARR61 pKa = 11.84EE62 pKa = 4.33LLRR65 pKa = 11.84YY66 pKa = 6.36WCRR69 pKa = 11.84RR70 pKa = 11.84EE71 pKa = 3.94PVGDD75 pKa = 3.37TDD77 pKa = 4.91GVGKK81 pKa = 10.44SLFQIDD87 pKa = 3.8HH88 pKa = 6.66SSLEE92 pKa = 4.08KK93 pKa = 9.85QVRR96 pKa = 11.84RR97 pKa = 11.84SSAGGVEE104 pKa = 4.34RR105 pKa = 11.84EE106 pKa = 4.12
MM1 pKa = 7.92RR2 pKa = 11.84FGSDD6 pKa = 2.96RR7 pKa = 11.84SPSVAAVADD16 pKa = 4.12GARR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84PRR23 pKa = 11.84PRR25 pKa = 11.84ATDD28 pKa = 3.37EE29 pKa = 4.6GNGDD33 pKa = 3.54PVRR36 pKa = 11.84RR37 pKa = 11.84TGVCSSDD44 pKa = 2.98GVTPQKK50 pKa = 10.2WPPQRR55 pKa = 11.84NSKK58 pKa = 9.87LARR61 pKa = 11.84EE62 pKa = 4.33LLRR65 pKa = 11.84YY66 pKa = 6.36WCRR69 pKa = 11.84RR70 pKa = 11.84EE71 pKa = 3.94PVGDD75 pKa = 3.37TDD77 pKa = 4.91GVGKK81 pKa = 10.44SLFQIDD87 pKa = 3.8HH88 pKa = 6.66SSLEE92 pKa = 4.08KK93 pKa = 9.85QVRR96 pKa = 11.84RR97 pKa = 11.84SSAGGVEE104 pKa = 4.34RR105 pKa = 11.84EE106 pKa = 4.12
Molecular weight: 11.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1077767 |
44 |
2528 |
297.2 |
32.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.602 ± 0.049 | 0.74 ± 0.014 |
8.84 ± 0.055 | 9.275 ± 0.058 |
3.197 ± 0.028 | 8.346 ± 0.036 |
2.032 ± 0.021 | 4.252 ± 0.033 |
1.58 ± 0.026 | 8.954 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.661 ± 0.02 | 2.11 ± 0.022 |
4.71 ± 0.028 | 2.455 ± 0.023 |
6.48 ± 0.044 | 5.435 ± 0.034 |
6.416 ± 0.033 | 9.175 ± 0.039 |
1.1 ± 0.017 | 2.641 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
