
Capybara microvirus Cap3_SP_612
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
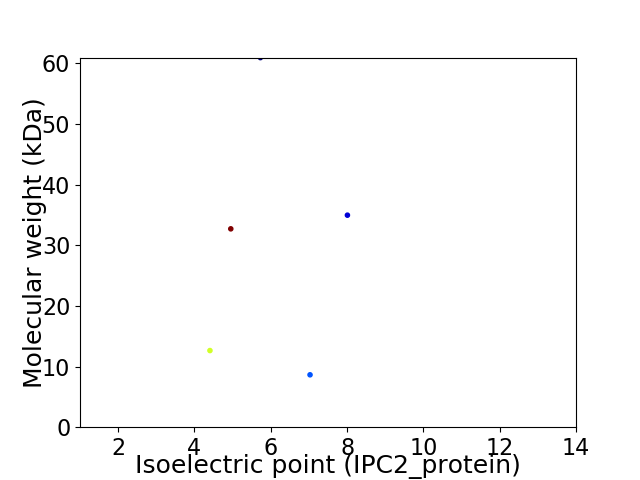
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
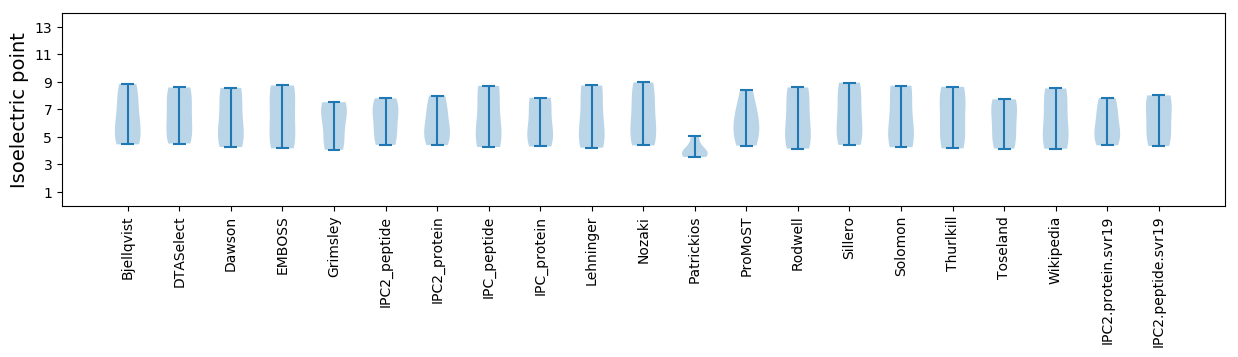
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W689|A0A4P8W689_9VIRU Replication initiator protein OS=Capybara microvirus Cap3_SP_612 OX=2585482 PE=4 SV=1
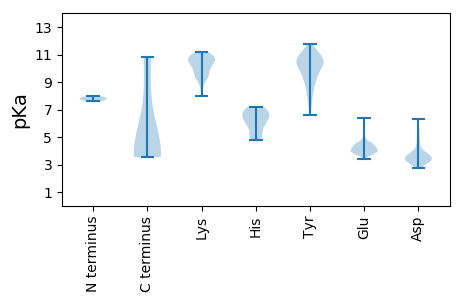
MM1 pKa = 7.73GILGVFRR8 pKa = 11.84TSSKK12 pKa = 10.86SRR14 pKa = 11.84IGEE17 pKa = 3.77INYY20 pKa = 9.34APSMTEE26 pKa = 3.78DD27 pKa = 3.07AHH29 pKa = 6.93LLSIEE34 pKa = 4.41QIVKK38 pKa = 10.22DD39 pKa = 3.64YY40 pKa = 11.33SRR42 pKa = 11.84GILHH46 pKa = 7.18PEE48 pKa = 4.01FNHH51 pKa = 6.4SAVYY55 pKa = 10.59DD56 pKa = 4.12DD57 pKa = 5.5GEE59 pKa = 4.54EE60 pKa = 3.85ISNVQNYY67 pKa = 10.44DD68 pKa = 3.48DD69 pKa = 4.59LTDD72 pKa = 4.41LFPNGVNSIYY82 pKa = 10.32HH83 pKa = 5.39LQDD86 pKa = 3.57RR87 pKa = 11.84EE88 pKa = 4.18PSSEE92 pKa = 4.0PQKK95 pKa = 10.84MNDD98 pKa = 3.38SAPEE102 pKa = 3.93SEE104 pKa = 4.67TNIEE108 pKa = 4.38TKK110 pKa = 10.91NEE112 pKa = 3.78
MM1 pKa = 7.73GILGVFRR8 pKa = 11.84TSSKK12 pKa = 10.86SRR14 pKa = 11.84IGEE17 pKa = 3.77INYY20 pKa = 9.34APSMTEE26 pKa = 3.78DD27 pKa = 3.07AHH29 pKa = 6.93LLSIEE34 pKa = 4.41QIVKK38 pKa = 10.22DD39 pKa = 3.64YY40 pKa = 11.33SRR42 pKa = 11.84GILHH46 pKa = 7.18PEE48 pKa = 4.01FNHH51 pKa = 6.4SAVYY55 pKa = 10.59DD56 pKa = 4.12DD57 pKa = 5.5GEE59 pKa = 4.54EE60 pKa = 3.85ISNVQNYY67 pKa = 10.44DD68 pKa = 3.48DD69 pKa = 4.59LTDD72 pKa = 4.41LFPNGVNSIYY82 pKa = 10.32HH83 pKa = 5.39LQDD86 pKa = 3.57RR87 pKa = 11.84EE88 pKa = 4.18PSSEE92 pKa = 4.0PQKK95 pKa = 10.84MNDD98 pKa = 3.38SAPEE102 pKa = 3.93SEE104 pKa = 4.67TNIEE108 pKa = 4.38TKK110 pKa = 10.91NEE112 pKa = 3.78
Molecular weight: 12.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W6G3|A0A4P8W6G3_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_612 OX=2585482 PE=3 SV=1
MM1 pKa = 7.78DD2 pKa = 6.31CIRR5 pKa = 11.84PFVKK9 pKa = 10.06TIGHH13 pKa = 6.48GLDD16 pKa = 3.35ANYY19 pKa = 10.13IPLPCGRR26 pKa = 11.84CLACRR31 pKa = 11.84VNYY34 pKa = 7.83TRR36 pKa = 11.84QWTQRR41 pKa = 11.84LLAEE45 pKa = 4.87SYY47 pKa = 10.23HH48 pKa = 6.33SKK50 pKa = 10.41NAYY53 pKa = 9.97FLTLTYY59 pKa = 10.62EE60 pKa = 4.41DD61 pKa = 3.82SCLPRR66 pKa = 11.84DD67 pKa = 4.7DD68 pKa = 5.2NDD70 pKa = 3.6NPCVCKK76 pKa = 10.3RR77 pKa = 11.84DD78 pKa = 3.12IQLFIKK84 pKa = 10.28RR85 pKa = 11.84LRR87 pKa = 11.84KK88 pKa = 9.56HH89 pKa = 6.31YY90 pKa = 9.24PDD92 pKa = 3.41SNIRR96 pKa = 11.84YY97 pKa = 8.58FLGSEE102 pKa = 4.15YY103 pKa = 11.03GEE105 pKa = 4.09EE106 pKa = 4.12TLRR109 pKa = 11.84PHH111 pKa = 5.56YY112 pKa = 10.23HH113 pKa = 6.81AIFYY117 pKa = 9.19NLPDD121 pKa = 3.61EE122 pKa = 6.35LIFQPQSAGGDD133 pKa = 3.01QWHH136 pKa = 5.97YY137 pKa = 10.72HH138 pKa = 6.91DD139 pKa = 5.6KK140 pKa = 9.44SQHH143 pKa = 5.11MINRR147 pKa = 11.84KK148 pKa = 9.49INDD151 pKa = 3.18YY152 pKa = 7.27WQKK155 pKa = 10.92GFIEE159 pKa = 4.29IDD161 pKa = 3.69PINRR165 pKa = 11.84QRR167 pKa = 11.84AEE169 pKa = 3.79YY170 pKa = 8.22CAKK173 pKa = 10.31YY174 pKa = 10.01FVFRR178 pKa = 11.84KK179 pKa = 9.97DD180 pKa = 3.22VEE182 pKa = 4.45QILVPNFNLMSRR194 pKa = 11.84KK195 pKa = 9.47PGIGYY200 pKa = 8.72QRR202 pKa = 11.84IDD204 pKa = 3.76EE205 pKa = 4.6IKK207 pKa = 10.97DD208 pKa = 2.99KK209 pKa = 11.03VRR211 pKa = 11.84YY212 pKa = 9.27YY213 pKa = 11.0NLNGMLTDD221 pKa = 3.49KK222 pKa = 10.31GTYY225 pKa = 9.21CAIPRR230 pKa = 11.84YY231 pKa = 9.3YY232 pKa = 10.27KK233 pKa = 10.7DD234 pKa = 3.26KK235 pKa = 10.59IYY237 pKa = 10.94SKK239 pKa = 9.86EE240 pKa = 3.82EE241 pKa = 3.68RR242 pKa = 11.84EE243 pKa = 4.11EE244 pKa = 3.94LFNRR248 pKa = 11.84YY249 pKa = 9.67LNMRR253 pKa = 11.84CTLSIEE259 pKa = 4.13EE260 pKa = 4.53IEE262 pKa = 4.51FNRR265 pKa = 11.84QKK267 pKa = 11.08LQGDD271 pKa = 3.92EE272 pKa = 4.69LEE274 pKa = 4.48EE275 pKa = 3.95SLIRR279 pKa = 11.84NMTFKK284 pKa = 10.84NKK286 pKa = 9.06KK287 pKa = 9.44RR288 pKa = 11.84KK289 pKa = 9.18LL290 pKa = 3.57
MM1 pKa = 7.78DD2 pKa = 6.31CIRR5 pKa = 11.84PFVKK9 pKa = 10.06TIGHH13 pKa = 6.48GLDD16 pKa = 3.35ANYY19 pKa = 10.13IPLPCGRR26 pKa = 11.84CLACRR31 pKa = 11.84VNYY34 pKa = 7.83TRR36 pKa = 11.84QWTQRR41 pKa = 11.84LLAEE45 pKa = 4.87SYY47 pKa = 10.23HH48 pKa = 6.33SKK50 pKa = 10.41NAYY53 pKa = 9.97FLTLTYY59 pKa = 10.62EE60 pKa = 4.41DD61 pKa = 3.82SCLPRR66 pKa = 11.84DD67 pKa = 4.7DD68 pKa = 5.2NDD70 pKa = 3.6NPCVCKK76 pKa = 10.3RR77 pKa = 11.84DD78 pKa = 3.12IQLFIKK84 pKa = 10.28RR85 pKa = 11.84LRR87 pKa = 11.84KK88 pKa = 9.56HH89 pKa = 6.31YY90 pKa = 9.24PDD92 pKa = 3.41SNIRR96 pKa = 11.84YY97 pKa = 8.58FLGSEE102 pKa = 4.15YY103 pKa = 11.03GEE105 pKa = 4.09EE106 pKa = 4.12TLRR109 pKa = 11.84PHH111 pKa = 5.56YY112 pKa = 10.23HH113 pKa = 6.81AIFYY117 pKa = 9.19NLPDD121 pKa = 3.61EE122 pKa = 6.35LIFQPQSAGGDD133 pKa = 3.01QWHH136 pKa = 5.97YY137 pKa = 10.72HH138 pKa = 6.91DD139 pKa = 5.6KK140 pKa = 9.44SQHH143 pKa = 5.11MINRR147 pKa = 11.84KK148 pKa = 9.49INDD151 pKa = 3.18YY152 pKa = 7.27WQKK155 pKa = 10.92GFIEE159 pKa = 4.29IDD161 pKa = 3.69PINRR165 pKa = 11.84QRR167 pKa = 11.84AEE169 pKa = 3.79YY170 pKa = 8.22CAKK173 pKa = 10.31YY174 pKa = 10.01FVFRR178 pKa = 11.84KK179 pKa = 9.97DD180 pKa = 3.22VEE182 pKa = 4.45QILVPNFNLMSRR194 pKa = 11.84KK195 pKa = 9.47PGIGYY200 pKa = 8.72QRR202 pKa = 11.84IDD204 pKa = 3.76EE205 pKa = 4.6IKK207 pKa = 10.97DD208 pKa = 2.99KK209 pKa = 11.03VRR211 pKa = 11.84YY212 pKa = 9.27YY213 pKa = 11.0NLNGMLTDD221 pKa = 3.49KK222 pKa = 10.31GTYY225 pKa = 9.21CAIPRR230 pKa = 11.84YY231 pKa = 9.3YY232 pKa = 10.27KK233 pKa = 10.7DD234 pKa = 3.26KK235 pKa = 10.59IYY237 pKa = 10.94SKK239 pKa = 9.86EE240 pKa = 3.82EE241 pKa = 3.68RR242 pKa = 11.84EE243 pKa = 4.11EE244 pKa = 3.94LFNRR248 pKa = 11.84YY249 pKa = 9.67LNMRR253 pKa = 11.84CTLSIEE259 pKa = 4.13EE260 pKa = 4.53IEE262 pKa = 4.51FNRR265 pKa = 11.84QKK267 pKa = 11.08LQGDD271 pKa = 3.92EE272 pKa = 4.69LEE274 pKa = 4.48EE275 pKa = 3.95SLIRR279 pKa = 11.84NMTFKK284 pKa = 10.84NKK286 pKa = 9.06KK287 pKa = 9.44RR288 pKa = 11.84KK289 pKa = 9.18LL290 pKa = 3.57
Molecular weight: 34.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1318 |
80 |
539 |
263.6 |
29.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.297 ± 1.151 | 1.29 ± 0.587 |
4.856 ± 0.614 | 7.056 ± 0.536 |
4.401 ± 0.772 | 6.222 ± 0.678 |
2.124 ± 0.363 | 6.753 ± 0.317 |
5.311 ± 0.737 | 7.891 ± 0.467 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.352 ± 0.152 | 7.891 ± 0.554 |
5.008 ± 0.461 | 4.021 ± 0.314 |
5.008 ± 0.943 | 6.601 ± 1.029 |
6.07 ± 1.123 | 4.552 ± 0.717 |
0.835 ± 0.116 | 5.463 ± 0.64 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
