
Pseudomonas phage Pf3 (Bacteriophage Pf3)
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; Tertilicivirus; Pseudomonas virus Pf3
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
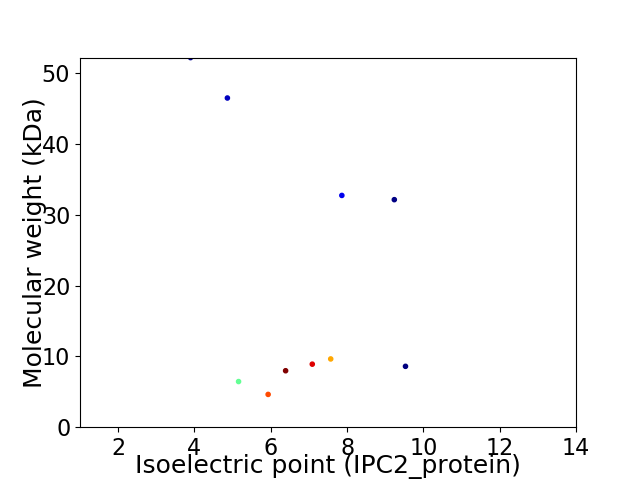
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P03625|G6P_BPPF3 Head virion protein G6P OS=Pseudomonas phage Pf3 OX=10872 GN=VI PE=3 SV=1
MM1 pKa = 7.41GLHH4 pKa = 6.01YY5 pKa = 11.01LFGFCLALFSFSAIAAGPVSTEE27 pKa = 3.79VAAGTTTYY35 pKa = 10.75RR36 pKa = 11.84VTNTTVRR43 pKa = 11.84TPPNVTLSPVRR54 pKa = 11.84DD55 pKa = 3.3ITPYY59 pKa = 10.38VEE61 pKa = 4.99KK62 pKa = 10.58IPNKK66 pKa = 10.25GLAQAAQGRR75 pKa = 11.84LIVAQRR81 pKa = 11.84AASVPVTGFFNVSGAVVKK99 pKa = 10.88SGAKK103 pKa = 9.83SFLRR107 pKa = 11.84SAGRR111 pKa = 11.84ASGIGLGLAALLEE124 pKa = 4.38AADD127 pKa = 3.63WVFDD131 pKa = 3.88EE132 pKa = 5.02EE133 pKa = 4.9GEE135 pKa = 4.35IVKK138 pKa = 9.23PLPGGGSPVLMPRR151 pKa = 11.84PVILNEE157 pKa = 3.81YY158 pKa = 8.63TVTGSAGQWSISKK171 pKa = 10.01EE172 pKa = 4.01YY173 pKa = 10.64EE174 pKa = 3.66PDD176 pKa = 3.49PRR178 pKa = 11.84SVPGWYY184 pKa = 9.59SYY186 pKa = 11.22NGNPVWVSAVEE197 pKa = 4.26DD198 pKa = 3.56VGFTWRR204 pKa = 11.84YY205 pKa = 7.74WYY207 pKa = 10.32FADD210 pKa = 5.03VLMDD214 pKa = 3.55GQGRR218 pKa = 11.84PNYY221 pKa = 9.67LVAYY225 pKa = 9.32SDD227 pKa = 3.91SGPNEE232 pKa = 3.66YY233 pKa = 9.25WQDD236 pKa = 3.27VGGYY240 pKa = 9.57SLDD243 pKa = 3.76SLPTEE248 pKa = 4.31PEE250 pKa = 4.11FVPLTDD256 pKa = 4.23AEE258 pKa = 4.42LEE260 pKa = 4.15AGIDD264 pKa = 3.39QYY266 pKa = 11.69YY267 pKa = 10.45EE268 pKa = 4.38PDD270 pKa = 3.97PDD272 pKa = 3.15DD273 pKa = 3.43WRR275 pKa = 11.84NLFPYY280 pKa = 10.0IEE282 pKa = 5.08PDD284 pKa = 2.96SFTIEE289 pKa = 4.13TPIPSLDD296 pKa = 3.53LSPVVSSSTNNQTGKK311 pKa = 8.66VTVTEE316 pKa = 4.43TTTSVDD322 pKa = 4.82FEE324 pKa = 4.64VSDD327 pKa = 4.29NNSSQPSISVNEE339 pKa = 4.15TTTEE343 pKa = 3.75NVYY346 pKa = 10.89VDD348 pKa = 3.76GDD350 pKa = 4.16LVSSEE355 pKa = 4.39TNTTVTNPPSSGTSTPPSSGSGSDD379 pKa = 3.46FQLPSFCSWATAVCDD394 pKa = 3.54WFDD397 pKa = 3.41WTQEE401 pKa = 4.48PIDD404 pKa = 4.64EE405 pKa = 4.66EE406 pKa = 5.02PDD408 pKa = 3.17LSGIISDD415 pKa = 3.69IDD417 pKa = 3.41DD418 pKa = 4.04LEE420 pKa = 4.23RR421 pKa = 11.84TKK423 pKa = 10.89DD424 pKa = 3.19ISFGSKK430 pKa = 9.61SCPAPIALDD439 pKa = 3.43IEE441 pKa = 4.39FLDD444 pKa = 3.73MSVDD448 pKa = 4.61LSFEE452 pKa = 4.31WFCEE456 pKa = 3.79LAGIIYY462 pKa = 9.03FMVMASAYY470 pKa = 10.25VLAAYY475 pKa = 7.31ITLGVVRR482 pKa = 11.84GG483 pKa = 3.72
MM1 pKa = 7.41GLHH4 pKa = 6.01YY5 pKa = 11.01LFGFCLALFSFSAIAAGPVSTEE27 pKa = 3.79VAAGTTTYY35 pKa = 10.75RR36 pKa = 11.84VTNTTVRR43 pKa = 11.84TPPNVTLSPVRR54 pKa = 11.84DD55 pKa = 3.3ITPYY59 pKa = 10.38VEE61 pKa = 4.99KK62 pKa = 10.58IPNKK66 pKa = 10.25GLAQAAQGRR75 pKa = 11.84LIVAQRR81 pKa = 11.84AASVPVTGFFNVSGAVVKK99 pKa = 10.88SGAKK103 pKa = 9.83SFLRR107 pKa = 11.84SAGRR111 pKa = 11.84ASGIGLGLAALLEE124 pKa = 4.38AADD127 pKa = 3.63WVFDD131 pKa = 3.88EE132 pKa = 5.02EE133 pKa = 4.9GEE135 pKa = 4.35IVKK138 pKa = 9.23PLPGGGSPVLMPRR151 pKa = 11.84PVILNEE157 pKa = 3.81YY158 pKa = 8.63TVTGSAGQWSISKK171 pKa = 10.01EE172 pKa = 4.01YY173 pKa = 10.64EE174 pKa = 3.66PDD176 pKa = 3.49PRR178 pKa = 11.84SVPGWYY184 pKa = 9.59SYY186 pKa = 11.22NGNPVWVSAVEE197 pKa = 4.26DD198 pKa = 3.56VGFTWRR204 pKa = 11.84YY205 pKa = 7.74WYY207 pKa = 10.32FADD210 pKa = 5.03VLMDD214 pKa = 3.55GQGRR218 pKa = 11.84PNYY221 pKa = 9.67LVAYY225 pKa = 9.32SDD227 pKa = 3.91SGPNEE232 pKa = 3.66YY233 pKa = 9.25WQDD236 pKa = 3.27VGGYY240 pKa = 9.57SLDD243 pKa = 3.76SLPTEE248 pKa = 4.31PEE250 pKa = 4.11FVPLTDD256 pKa = 4.23AEE258 pKa = 4.42LEE260 pKa = 4.15AGIDD264 pKa = 3.39QYY266 pKa = 11.69YY267 pKa = 10.45EE268 pKa = 4.38PDD270 pKa = 3.97PDD272 pKa = 3.15DD273 pKa = 3.43WRR275 pKa = 11.84NLFPYY280 pKa = 10.0IEE282 pKa = 5.08PDD284 pKa = 2.96SFTIEE289 pKa = 4.13TPIPSLDD296 pKa = 3.53LSPVVSSSTNNQTGKK311 pKa = 8.66VTVTEE316 pKa = 4.43TTTSVDD322 pKa = 4.82FEE324 pKa = 4.64VSDD327 pKa = 4.29NNSSQPSISVNEE339 pKa = 4.15TTTEE343 pKa = 3.75NVYY346 pKa = 10.89VDD348 pKa = 3.76GDD350 pKa = 4.16LVSSEE355 pKa = 4.39TNTTVTNPPSSGTSTPPSSGSGSDD379 pKa = 3.46FQLPSFCSWATAVCDD394 pKa = 3.54WFDD397 pKa = 3.41WTQEE401 pKa = 4.48PIDD404 pKa = 4.64EE405 pKa = 4.66EE406 pKa = 5.02PDD408 pKa = 3.17LSGIISDD415 pKa = 3.69IDD417 pKa = 3.41DD418 pKa = 4.04LEE420 pKa = 4.23RR421 pKa = 11.84TKK423 pKa = 10.89DD424 pKa = 3.19ISFGSKK430 pKa = 9.61SCPAPIALDD439 pKa = 3.43IEE441 pKa = 4.39FLDD444 pKa = 3.73MSVDD448 pKa = 4.61LSFEE452 pKa = 4.31WFCEE456 pKa = 3.79LAGIIYY462 pKa = 9.03FMVMASAYY470 pKa = 10.25VLAAYY475 pKa = 7.31ITLGVVRR482 pKa = 11.84GG483 pKa = 3.72
Molecular weight: 52.2 kDa
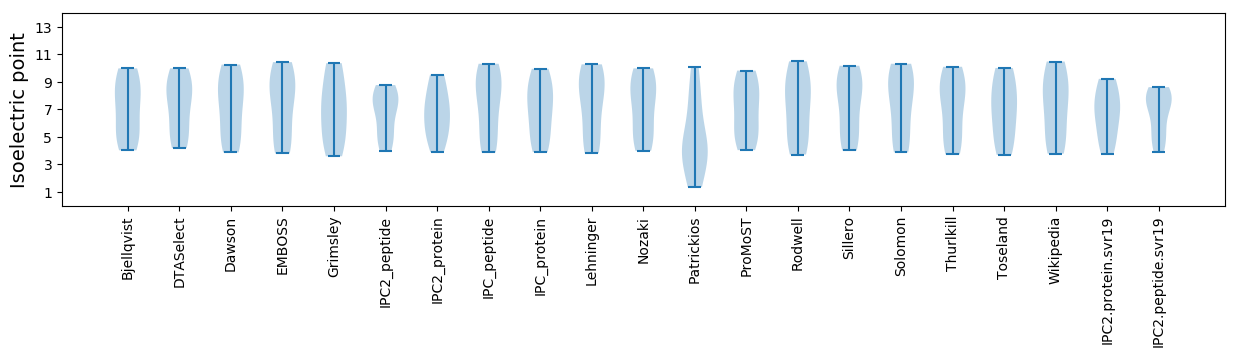
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P03627-2|REP-2_BPPF3 Isoform of P03627 Isoform G10P of Replication-associated protein G2P OS=Pseudomonas phage Pf3 OX=10872 GN=II PE=4 SV=1
MM1 pKa = 7.75GDD3 pKa = 2.89ATFYY7 pKa = 9.38QLRR10 pKa = 11.84RR11 pKa = 11.84IVKK14 pKa = 8.07RR15 pKa = 11.84WRR17 pKa = 11.84FADD20 pKa = 3.31ARR22 pKa = 11.84ASGSIHH28 pKa = 7.69RR29 pKa = 11.84IRR31 pKa = 11.84SAKK34 pKa = 10.11KK35 pKa = 7.82YY36 pKa = 9.23LQVPEE41 pKa = 4.14TLSRR45 pKa = 11.84VLGASEE51 pKa = 3.71WMPEE55 pKa = 3.79NEE57 pKa = 4.81LLSILYY63 pKa = 8.99FLKK66 pKa = 10.18SQGYY70 pKa = 8.89EE71 pKa = 3.97FLL73 pKa = 5.21
MM1 pKa = 7.75GDD3 pKa = 2.89ATFYY7 pKa = 9.38QLRR10 pKa = 11.84RR11 pKa = 11.84IVKK14 pKa = 8.07RR15 pKa = 11.84WRR17 pKa = 11.84FADD20 pKa = 3.31ARR22 pKa = 11.84ASGSIHH28 pKa = 7.69RR29 pKa = 11.84IRR31 pKa = 11.84SAKK34 pKa = 10.11KK35 pKa = 7.82YY36 pKa = 9.23LQVPEE41 pKa = 4.14TLSRR45 pKa = 11.84VLGASEE51 pKa = 3.71WMPEE55 pKa = 3.79NEE57 pKa = 4.81LLSILYY63 pKa = 8.99FLKK66 pKa = 10.18SQGYY70 pKa = 8.89EE71 pKa = 3.97FLL73 pKa = 5.21
Molecular weight: 8.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1909 |
44 |
483 |
190.9 |
20.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.539 ± 0.695 | 1.257 ± 0.295 |
5.186 ± 0.658 | 4.872 ± 0.525 |
4.819 ± 0.422 | 6.967 ± 0.377 |
2.095 ± 0.692 | 5.395 ± 0.539 |
3.248 ± 0.572 | 9.953 ± 1.385 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.938 ± 0.231 | 3.248 ± 0.332 |
5.605 ± 0.766 | 3.353 ± 0.417 |
5.186 ± 1.058 | 9.115 ± 0.765 |
5.605 ± 0.695 | 8.853 ± 0.779 |
1.886 ± 0.477 | 2.881 ± 0.558 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
