
Firmicutes bacterium CAG:114
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; environmental samples
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
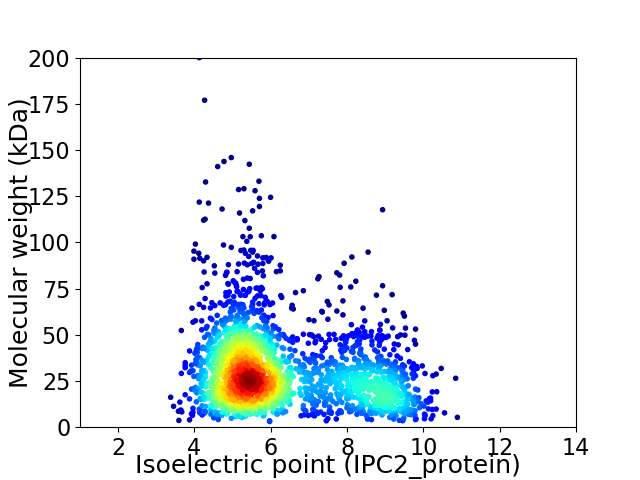
Virtual 2D-PAGE plot for 2222 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5HG04|R5HG04_9FIRM Uncharacterized protein OS=Firmicutes bacterium CAG:114 OX=1263001 GN=BN469_01834 PE=4 SV=1
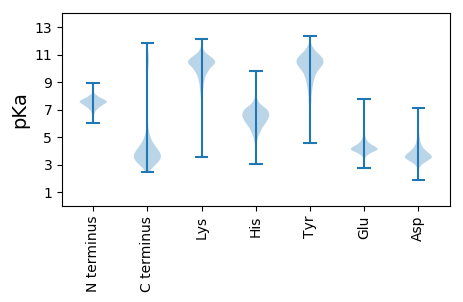
MM1 pKa = 7.62KK2 pKa = 10.56KK3 pKa = 9.21MLALTLALVMALGLCACGGDD23 pKa = 3.62TTDD26 pKa = 5.03NGDD29 pKa = 3.79APAEE33 pKa = 4.05NPEE36 pKa = 4.37TNSPAEE42 pKa = 4.28TVTLDD47 pKa = 3.72LPPVEE52 pKa = 5.6DD53 pKa = 3.76LTGTDD58 pKa = 3.23TSAIAGVEE66 pKa = 4.04DD67 pKa = 4.2GVLTVAMEE75 pKa = 4.34CTYY78 pKa = 11.43APYY81 pKa = 10.94NWTQMDD87 pKa = 4.23DD88 pKa = 3.66SNGAVPIKK96 pKa = 10.62GSSEE100 pKa = 3.94YY101 pKa = 11.38ANGYY105 pKa = 10.54DD106 pKa = 3.6VMIAKK111 pKa = 9.28RR112 pKa = 11.84ICEE115 pKa = 3.9AYY117 pKa = 9.46GWEE120 pKa = 4.09LEE122 pKa = 4.72IVRR125 pKa = 11.84TDD127 pKa = 3.11WNSLVPGIQSGLYY140 pKa = 9.25DD141 pKa = 3.87AVIAGQSMTEE151 pKa = 3.81DD152 pKa = 2.78RR153 pKa = 11.84MEE155 pKa = 4.11QVDD158 pKa = 4.07FAGPYY163 pKa = 8.1YY164 pKa = 9.88YY165 pKa = 11.17ASIVCVAKK173 pKa = 10.74DD174 pKa = 3.61GSPQASAKK182 pKa = 10.37GISEE186 pKa = 4.3LTGTATAQIEE196 pKa = 4.8TIWYY200 pKa = 6.97DD201 pKa = 3.21TCLPQIKK208 pKa = 9.81GAEE211 pKa = 4.0IQTAAEE217 pKa = 4.03TAPAMLMALEE227 pKa = 4.36TGAVEE232 pKa = 6.23FICTDD237 pKa = 2.7IPTAKK242 pKa = 10.1GAVAAYY248 pKa = 10.28DD249 pKa = 3.74DD250 pKa = 4.25MVILDD255 pKa = 3.94FTDD258 pKa = 3.53TEE260 pKa = 5.23DD261 pKa = 3.61NFQVDD266 pKa = 3.8EE267 pKa = 4.3GDD269 pKa = 3.48INIGISLKK277 pKa = 10.69KK278 pKa = 10.82GNTEE282 pKa = 4.59LKK284 pKa = 10.51DD285 pKa = 4.17AINKK289 pKa = 8.14VVSALNKK296 pKa = 10.77DD297 pKa = 3.6NFDD300 pKa = 3.7NLMDD304 pKa = 4.04YY305 pKa = 10.71AVSIQPLNQQ314 pKa = 3.08
MM1 pKa = 7.62KK2 pKa = 10.56KK3 pKa = 9.21MLALTLALVMALGLCACGGDD23 pKa = 3.62TTDD26 pKa = 5.03NGDD29 pKa = 3.79APAEE33 pKa = 4.05NPEE36 pKa = 4.37TNSPAEE42 pKa = 4.28TVTLDD47 pKa = 3.72LPPVEE52 pKa = 5.6DD53 pKa = 3.76LTGTDD58 pKa = 3.23TSAIAGVEE66 pKa = 4.04DD67 pKa = 4.2GVLTVAMEE75 pKa = 4.34CTYY78 pKa = 11.43APYY81 pKa = 10.94NWTQMDD87 pKa = 4.23DD88 pKa = 3.66SNGAVPIKK96 pKa = 10.62GSSEE100 pKa = 3.94YY101 pKa = 11.38ANGYY105 pKa = 10.54DD106 pKa = 3.6VMIAKK111 pKa = 9.28RR112 pKa = 11.84ICEE115 pKa = 3.9AYY117 pKa = 9.46GWEE120 pKa = 4.09LEE122 pKa = 4.72IVRR125 pKa = 11.84TDD127 pKa = 3.11WNSLVPGIQSGLYY140 pKa = 9.25DD141 pKa = 3.87AVIAGQSMTEE151 pKa = 3.81DD152 pKa = 2.78RR153 pKa = 11.84MEE155 pKa = 4.11QVDD158 pKa = 4.07FAGPYY163 pKa = 8.1YY164 pKa = 9.88YY165 pKa = 11.17ASIVCVAKK173 pKa = 10.74DD174 pKa = 3.61GSPQASAKK182 pKa = 10.37GISEE186 pKa = 4.3LTGTATAQIEE196 pKa = 4.8TIWYY200 pKa = 6.97DD201 pKa = 3.21TCLPQIKK208 pKa = 9.81GAEE211 pKa = 4.0IQTAAEE217 pKa = 4.03TAPAMLMALEE227 pKa = 4.36TGAVEE232 pKa = 6.23FICTDD237 pKa = 2.7IPTAKK242 pKa = 10.1GAVAAYY248 pKa = 10.28DD249 pKa = 3.74DD250 pKa = 4.25MVILDD255 pKa = 3.94FTDD258 pKa = 3.53TEE260 pKa = 5.23DD261 pKa = 3.61NFQVDD266 pKa = 3.8EE267 pKa = 4.3GDD269 pKa = 3.48INIGISLKK277 pKa = 10.69KK278 pKa = 10.82GNTEE282 pKa = 4.59LKK284 pKa = 10.51DD285 pKa = 4.17AINKK289 pKa = 8.14VVSALNKK296 pKa = 10.77DD297 pKa = 3.6NFDD300 pKa = 3.7NLMDD304 pKa = 4.04YY305 pKa = 10.71AVSIQPLNQQ314 pKa = 3.08
Molecular weight: 33.47 kDa
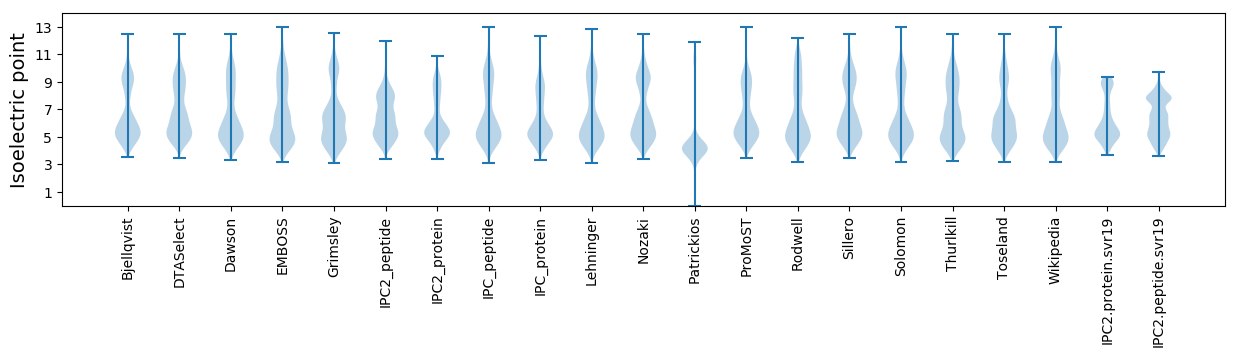
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5HBE7|R5HBE7_9FIRM Predicted transcriptional regulator OS=Firmicutes bacterium CAG:114 OX=1263001 GN=BN469_01744 PE=4 SV=1
MM1 pKa = 7.06VRR3 pKa = 11.84TYY5 pKa = 10.22QPKK8 pKa = 9.24KK9 pKa = 7.68RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.69EE15 pKa = 3.5HH16 pKa = 6.14GFRR19 pKa = 11.84KK20 pKa = 9.99RR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.58VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84ARR41 pKa = 11.84LTHH44 pKa = 6.31
MM1 pKa = 7.06VRR3 pKa = 11.84TYY5 pKa = 10.22QPKK8 pKa = 9.24KK9 pKa = 7.68RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.69EE15 pKa = 3.5HH16 pKa = 6.14GFRR19 pKa = 11.84KK20 pKa = 9.99RR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.58VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84ARR41 pKa = 11.84LTHH44 pKa = 6.31
Molecular weight: 5.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
642334 |
29 |
1845 |
289.1 |
31.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.542 ± 0.056 | 1.562 ± 0.024 |
5.178 ± 0.048 | 6.492 ± 0.054 |
3.744 ± 0.043 | 8.283 ± 0.067 |
1.983 ± 0.032 | 5.264 ± 0.049 |
4.395 ± 0.049 | 10.62 ± 0.083 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.616 ± 0.028 | 3.037 ± 0.036 |
4.499 ± 0.039 | 4.209 ± 0.039 |
5.726 ± 0.066 | 5.333 ± 0.042 |
5.805 ± 0.06 | 7.32 ± 0.043 |
1.102 ± 0.022 | 3.286 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
