
Gopherus associated circular DNA virus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.04
Get precalculated fractions of proteins
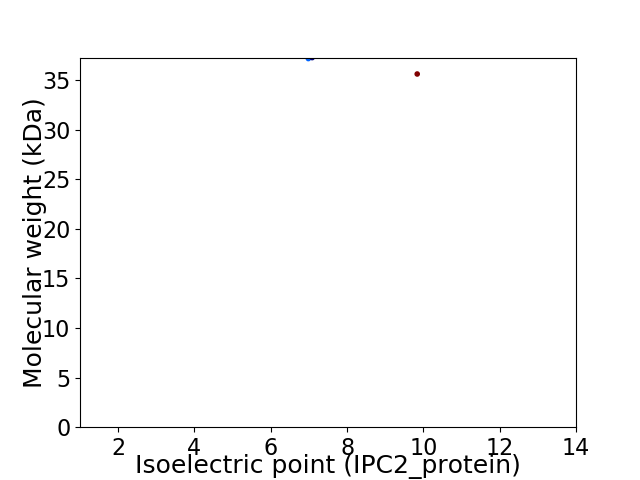
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A384WVX5|A0A384WVX5_9VIRU Replication associated protein OS=Gopherus associated circular DNA virus 1 OX=2041419 PE=3 SV=1
MM1 pKa = 7.27EE2 pKa = 4.18QTGEE6 pKa = 3.94PDD8 pKa = 3.63GNSSSVAQRR17 pKa = 11.84RR18 pKa = 11.84FHH20 pKa = 6.98LNAKK24 pKa = 7.72TLFLTYY30 pKa = 8.75PQCDD34 pKa = 3.78LPRR37 pKa = 11.84EE38 pKa = 4.2EE39 pKa = 5.28ALSLLEE45 pKa = 3.98AAFMTRR51 pKa = 11.84QRR53 pKa = 11.84RR54 pKa = 11.84IIEE57 pKa = 3.85YY58 pKa = 10.19LIASEE63 pKa = 4.15KK64 pKa = 10.38HH65 pKa = 6.19ADD67 pKa = 3.84GTPHH71 pKa = 6.64LHH73 pKa = 6.9CFVKK77 pKa = 10.64LNKK80 pKa = 9.78AVNLKK85 pKa = 10.54RR86 pKa = 11.84PTEE89 pKa = 3.98LDD91 pKa = 3.43LKK93 pKa = 9.64NHH95 pKa = 6.38HH96 pKa = 6.82GNYY99 pKa = 9.05QSCRR103 pKa = 11.84SPEE106 pKa = 3.63AVRR109 pKa = 11.84RR110 pKa = 11.84YY111 pKa = 9.37CSKK114 pKa = 11.03DD115 pKa = 3.11GNFLTNITSGTDD127 pKa = 3.26SQEE130 pKa = 4.29GPWRR134 pKa = 11.84KK135 pKa = 9.14ARR137 pKa = 11.84KK138 pKa = 8.64IVKK141 pKa = 9.94EE142 pKa = 4.09GGDD145 pKa = 3.57LKK147 pKa = 10.99EE148 pKa = 4.43ALLALEE154 pKa = 4.46EE155 pKa = 4.46TEE157 pKa = 3.99KK158 pKa = 10.2TARR161 pKa = 11.84DD162 pKa = 3.37LTIYY166 pKa = 10.73GSTISQALRR175 pKa = 11.84SLKK178 pKa = 10.31PVTEE182 pKa = 4.19LTSEE186 pKa = 4.08QTLTSYY192 pKa = 11.45GPLFQWDD199 pKa = 3.8QAKK202 pKa = 10.52HH203 pKa = 5.39SLLLFGNTEE212 pKa = 4.05TGKK215 pKa = 7.65TTLAKK220 pKa = 10.68LLIPKK225 pKa = 10.04ALFVRR230 pKa = 11.84HH231 pKa = 6.62LDD233 pKa = 3.6LLLSLTEE240 pKa = 3.87EE241 pKa = 4.21HH242 pKa = 6.79GGVILDD248 pKa = 4.31DD249 pKa = 3.46MTFKK253 pKa = 10.69HH254 pKa = 5.97LHH256 pKa = 6.94DD257 pKa = 4.22EE258 pKa = 4.23AQIALLDD265 pKa = 4.22LQDD268 pKa = 3.46EE269 pKa = 4.69THH271 pKa = 5.08IHH273 pKa = 3.89VRR275 pKa = 11.84YY276 pKa = 9.52RR277 pKa = 11.84VAVLPAKK284 pKa = 10.15LPRR287 pKa = 11.84IITSNKK293 pKa = 9.02EE294 pKa = 3.38PWEE297 pKa = 4.22VVNMANPAIARR308 pKa = 11.84RR309 pKa = 11.84VLCVRR314 pKa = 11.84WYY316 pKa = 10.41GWDD319 pKa = 3.37KK320 pKa = 11.03VPMFNVEE327 pKa = 3.67
MM1 pKa = 7.27EE2 pKa = 4.18QTGEE6 pKa = 3.94PDD8 pKa = 3.63GNSSSVAQRR17 pKa = 11.84RR18 pKa = 11.84FHH20 pKa = 6.98LNAKK24 pKa = 7.72TLFLTYY30 pKa = 8.75PQCDD34 pKa = 3.78LPRR37 pKa = 11.84EE38 pKa = 4.2EE39 pKa = 5.28ALSLLEE45 pKa = 3.98AAFMTRR51 pKa = 11.84QRR53 pKa = 11.84RR54 pKa = 11.84IIEE57 pKa = 3.85YY58 pKa = 10.19LIASEE63 pKa = 4.15KK64 pKa = 10.38HH65 pKa = 6.19ADD67 pKa = 3.84GTPHH71 pKa = 6.64LHH73 pKa = 6.9CFVKK77 pKa = 10.64LNKK80 pKa = 9.78AVNLKK85 pKa = 10.54RR86 pKa = 11.84PTEE89 pKa = 3.98LDD91 pKa = 3.43LKK93 pKa = 9.64NHH95 pKa = 6.38HH96 pKa = 6.82GNYY99 pKa = 9.05QSCRR103 pKa = 11.84SPEE106 pKa = 3.63AVRR109 pKa = 11.84RR110 pKa = 11.84YY111 pKa = 9.37CSKK114 pKa = 11.03DD115 pKa = 3.11GNFLTNITSGTDD127 pKa = 3.26SQEE130 pKa = 4.29GPWRR134 pKa = 11.84KK135 pKa = 9.14ARR137 pKa = 11.84KK138 pKa = 8.64IVKK141 pKa = 9.94EE142 pKa = 4.09GGDD145 pKa = 3.57LKK147 pKa = 10.99EE148 pKa = 4.43ALLALEE154 pKa = 4.46EE155 pKa = 4.46TEE157 pKa = 3.99KK158 pKa = 10.2TARR161 pKa = 11.84DD162 pKa = 3.37LTIYY166 pKa = 10.73GSTISQALRR175 pKa = 11.84SLKK178 pKa = 10.31PVTEE182 pKa = 4.19LTSEE186 pKa = 4.08QTLTSYY192 pKa = 11.45GPLFQWDD199 pKa = 3.8QAKK202 pKa = 10.52HH203 pKa = 5.39SLLLFGNTEE212 pKa = 4.05TGKK215 pKa = 7.65TTLAKK220 pKa = 10.68LLIPKK225 pKa = 10.04ALFVRR230 pKa = 11.84HH231 pKa = 6.62LDD233 pKa = 3.6LLLSLTEE240 pKa = 3.87EE241 pKa = 4.21HH242 pKa = 6.79GGVILDD248 pKa = 4.31DD249 pKa = 3.46MTFKK253 pKa = 10.69HH254 pKa = 5.97LHH256 pKa = 6.94DD257 pKa = 4.22EE258 pKa = 4.23AQIALLDD265 pKa = 4.22LQDD268 pKa = 3.46EE269 pKa = 4.69THH271 pKa = 5.08IHH273 pKa = 3.89VRR275 pKa = 11.84YY276 pKa = 9.52RR277 pKa = 11.84VAVLPAKK284 pKa = 10.15LPRR287 pKa = 11.84IITSNKK293 pKa = 9.02EE294 pKa = 3.38PWEE297 pKa = 4.22VVNMANPAIARR308 pKa = 11.84RR309 pKa = 11.84VLCVRR314 pKa = 11.84WYY316 pKa = 10.41GWDD319 pKa = 3.37KK320 pKa = 11.03VPMFNVEE327 pKa = 3.67
Molecular weight: 37.16 kDa
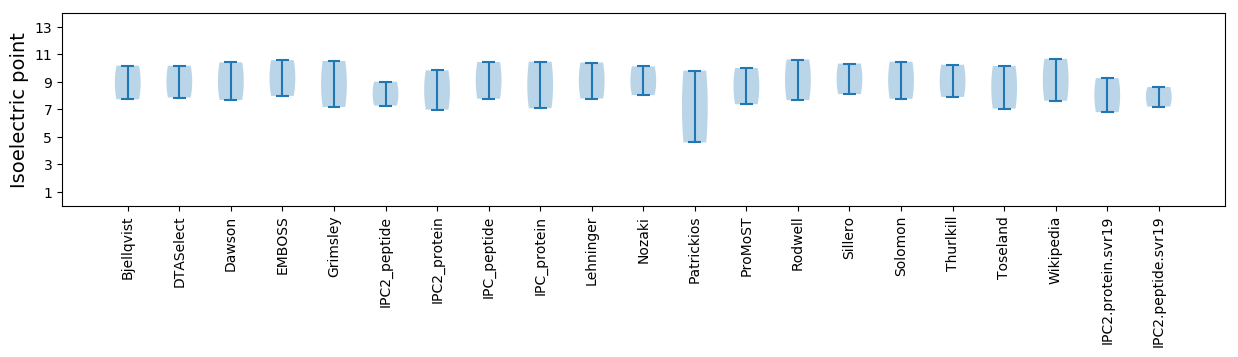
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A384WVX5|A0A384WVX5_9VIRU Replication associated protein OS=Gopherus associated circular DNA virus 1 OX=2041419 PE=3 SV=1
MM1 pKa = 7.13ARR3 pKa = 11.84SYY5 pKa = 10.76RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84TYY11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 9.21RR15 pKa = 11.84PTYY18 pKa = 8.83RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84INRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 8.36PMRR30 pKa = 11.84GRR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 8.43LKK36 pKa = 9.35QRR38 pKa = 11.84RR39 pKa = 11.84FVQRR43 pKa = 11.84NRR45 pKa = 11.84MRR47 pKa = 11.84AQIDD51 pKa = 3.61SNRR54 pKa = 11.84FSNNLGDD61 pKa = 3.62MSYY64 pKa = 7.73TTVRR68 pKa = 11.84DD69 pKa = 3.49THH71 pKa = 5.61WAVMQRR77 pKa = 11.84AVTYY81 pKa = 8.34GTDD84 pKa = 3.09AAGAPFNPKK93 pKa = 10.26AGTDD97 pKa = 3.17NQYY100 pKa = 11.22YY101 pKa = 9.88VGYY104 pKa = 8.96SLPVNYY110 pKa = 9.35FWSRR114 pKa = 11.84ITPGLRR120 pKa = 11.84HH121 pKa = 6.0YY122 pKa = 10.53YY123 pKa = 10.77NNYY126 pKa = 9.69KK127 pKa = 9.95YY128 pKa = 10.27ATVEE132 pKa = 3.78RR133 pKa = 11.84FLIKK137 pKa = 9.99ATFYY141 pKa = 11.24NSGSVLTKK149 pKa = 10.23EE150 pKa = 4.26VGILKK155 pKa = 9.94PDD157 pKa = 3.93AALGWPGINWTPTQLPSEE175 pKa = 4.27QPKK178 pKa = 10.37GVSSFITAHH187 pKa = 6.29PVAGSYY193 pKa = 7.65KK194 pKa = 8.55TLTMALTKK202 pKa = 10.4KK203 pKa = 10.6DD204 pKa = 3.51MYY206 pKa = 10.4QGSRR210 pKa = 11.84LEE212 pKa = 3.99PGRR215 pKa = 11.84GYY217 pKa = 10.78DD218 pKa = 4.64IISTGINGAGLLQPPEE234 pKa = 4.51YY235 pKa = 8.9PTFIYY240 pKa = 9.74IWQGNPIAGVEE251 pKa = 4.09AEE253 pKa = 4.35SFADD257 pKa = 3.81AVWVKK262 pKa = 6.92MTVWRR267 pKa = 11.84RR268 pKa = 11.84VKK270 pKa = 10.58FFGKK274 pKa = 8.84ATPLFAGRR282 pKa = 11.84TDD284 pKa = 3.47EE285 pKa = 4.3ATDD288 pKa = 3.55VYY290 pKa = 9.31TTGQFGTDD298 pKa = 3.27PGDD301 pKa = 3.76EE302 pKa = 3.98VDD304 pKa = 3.65EE305 pKa = 4.9AFTVEE310 pKa = 3.93
MM1 pKa = 7.13ARR3 pKa = 11.84SYY5 pKa = 10.76RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84TYY11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 9.21RR15 pKa = 11.84PTYY18 pKa = 8.83RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84INRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 8.36PMRR30 pKa = 11.84GRR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 8.43LKK36 pKa = 9.35QRR38 pKa = 11.84RR39 pKa = 11.84FVQRR43 pKa = 11.84NRR45 pKa = 11.84MRR47 pKa = 11.84AQIDD51 pKa = 3.61SNRR54 pKa = 11.84FSNNLGDD61 pKa = 3.62MSYY64 pKa = 7.73TTVRR68 pKa = 11.84DD69 pKa = 3.49THH71 pKa = 5.61WAVMQRR77 pKa = 11.84AVTYY81 pKa = 8.34GTDD84 pKa = 3.09AAGAPFNPKK93 pKa = 10.26AGTDD97 pKa = 3.17NQYY100 pKa = 11.22YY101 pKa = 9.88VGYY104 pKa = 8.96SLPVNYY110 pKa = 9.35FWSRR114 pKa = 11.84ITPGLRR120 pKa = 11.84HH121 pKa = 6.0YY122 pKa = 10.53YY123 pKa = 10.77NNYY126 pKa = 9.69KK127 pKa = 9.95YY128 pKa = 10.27ATVEE132 pKa = 3.78RR133 pKa = 11.84FLIKK137 pKa = 9.99ATFYY141 pKa = 11.24NSGSVLTKK149 pKa = 10.23EE150 pKa = 4.26VGILKK155 pKa = 9.94PDD157 pKa = 3.93AALGWPGINWTPTQLPSEE175 pKa = 4.27QPKK178 pKa = 10.37GVSSFITAHH187 pKa = 6.29PVAGSYY193 pKa = 7.65KK194 pKa = 8.55TLTMALTKK202 pKa = 10.4KK203 pKa = 10.6DD204 pKa = 3.51MYY206 pKa = 10.4QGSRR210 pKa = 11.84LEE212 pKa = 3.99PGRR215 pKa = 11.84GYY217 pKa = 10.78DD218 pKa = 4.64IISTGINGAGLLQPPEE234 pKa = 4.51YY235 pKa = 8.9PTFIYY240 pKa = 9.74IWQGNPIAGVEE251 pKa = 4.09AEE253 pKa = 4.35SFADD257 pKa = 3.81AVWVKK262 pKa = 6.92MTVWRR267 pKa = 11.84RR268 pKa = 11.84VKK270 pKa = 10.58FFGKK274 pKa = 8.84ATPLFAGRR282 pKa = 11.84TDD284 pKa = 3.47EE285 pKa = 4.3ATDD288 pKa = 3.55VYY290 pKa = 9.31TTGQFGTDD298 pKa = 3.27PGDD301 pKa = 3.76EE302 pKa = 3.98VDD304 pKa = 3.65EE305 pKa = 4.9AFTVEE310 pKa = 3.93
Molecular weight: 35.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
637 |
310 |
327 |
318.5 |
36.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.535 ± 0.138 | 0.785 ± 0.524 |
4.71 ± 0.129 | 5.651 ± 1.404 |
3.768 ± 0.5 | 6.593 ± 1.197 |
2.512 ± 1.031 | 4.239 ± 0.03 |
5.651 ± 0.543 | 9.262 ± 2.953 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.041 ± 0.36 | 4.239 ± 0.185 |
5.338 ± 0.528 | 3.611 ± 0.042 |
8.477 ± 1.447 | 5.181 ± 0.228 |
8.32 ± 0.475 | 5.651 ± 0.319 |
1.884 ± 0.25 | 4.553 ± 1.483 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
