
Capybara microvirus Cap3_SP_445
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.69
Get precalculated fractions of proteins
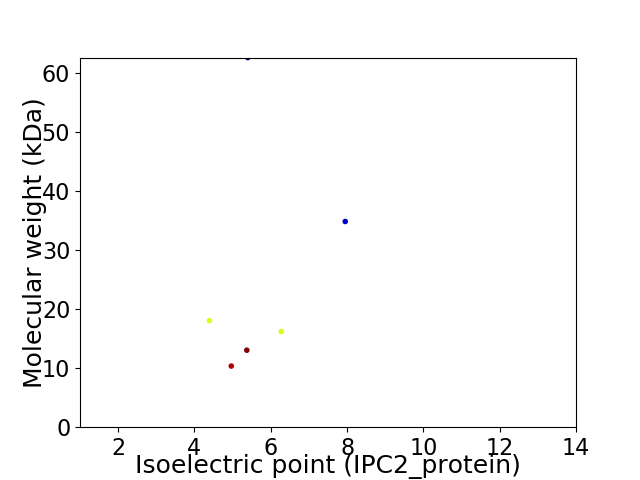
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W8L8|A0A4P8W8L8_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_445 OX=2585457 PE=3 SV=1
MM1 pKa = 7.17KK2 pKa = 10.3QNPSNCFKK10 pKa = 10.81ARR12 pKa = 11.84SDD14 pKa = 3.33GKK16 pKa = 10.38VFPGEE21 pKa = 4.05VFSPIVKK28 pKa = 9.8EE29 pKa = 3.86YY30 pKa = 11.29SLIDD34 pKa = 3.71SVIMIGDD41 pKa = 4.04TPRR44 pKa = 11.84DD45 pKa = 4.01FIIHH49 pKa = 5.87KK50 pKa = 8.89TPEE53 pKa = 4.91LINEE57 pKa = 3.88YY58 pKa = 10.83DD59 pKa = 3.3IDD61 pKa = 3.91KK62 pKa = 11.04QIAAEE67 pKa = 4.19AASCDD72 pKa = 3.33IKK74 pKa = 11.31SQILAAMQNGSIDD87 pKa = 3.54SLAWDD92 pKa = 4.9GSSGAIDD99 pKa = 3.83ATSMPSNLMEE109 pKa = 4.27AQNIINAGLNAYY121 pKa = 10.01DD122 pKa = 5.39SLDD125 pKa = 3.84PDD127 pKa = 4.17LKK129 pKa = 11.11KK130 pKa = 10.71KK131 pKa = 10.75GLDD134 pKa = 3.63FIKK137 pKa = 11.0NMTQTEE143 pKa = 4.41FEE145 pKa = 4.66SYY147 pKa = 9.75IKK149 pKa = 10.56SLIPSQPIEE158 pKa = 4.17KK159 pKa = 10.56DD160 pKa = 2.94GDD162 pKa = 3.77INGG165 pKa = 3.74
MM1 pKa = 7.17KK2 pKa = 10.3QNPSNCFKK10 pKa = 10.81ARR12 pKa = 11.84SDD14 pKa = 3.33GKK16 pKa = 10.38VFPGEE21 pKa = 4.05VFSPIVKK28 pKa = 9.8EE29 pKa = 3.86YY30 pKa = 11.29SLIDD34 pKa = 3.71SVIMIGDD41 pKa = 4.04TPRR44 pKa = 11.84DD45 pKa = 4.01FIIHH49 pKa = 5.87KK50 pKa = 8.89TPEE53 pKa = 4.91LINEE57 pKa = 3.88YY58 pKa = 10.83DD59 pKa = 3.3IDD61 pKa = 3.91KK62 pKa = 11.04QIAAEE67 pKa = 4.19AASCDD72 pKa = 3.33IKK74 pKa = 11.31SQILAAMQNGSIDD87 pKa = 3.54SLAWDD92 pKa = 4.9GSSGAIDD99 pKa = 3.83ATSMPSNLMEE109 pKa = 4.27AQNIINAGLNAYY121 pKa = 10.01DD122 pKa = 5.39SLDD125 pKa = 3.84PDD127 pKa = 4.17LKK129 pKa = 11.11KK130 pKa = 10.71KK131 pKa = 10.75GLDD134 pKa = 3.63FIKK137 pKa = 11.0NMTQTEE143 pKa = 4.41FEE145 pKa = 4.66SYY147 pKa = 9.75IKK149 pKa = 10.56SLIPSQPIEE158 pKa = 4.17KK159 pKa = 10.56DD160 pKa = 2.94GDD162 pKa = 3.77INGG165 pKa = 3.74
Molecular weight: 18.04 kDa
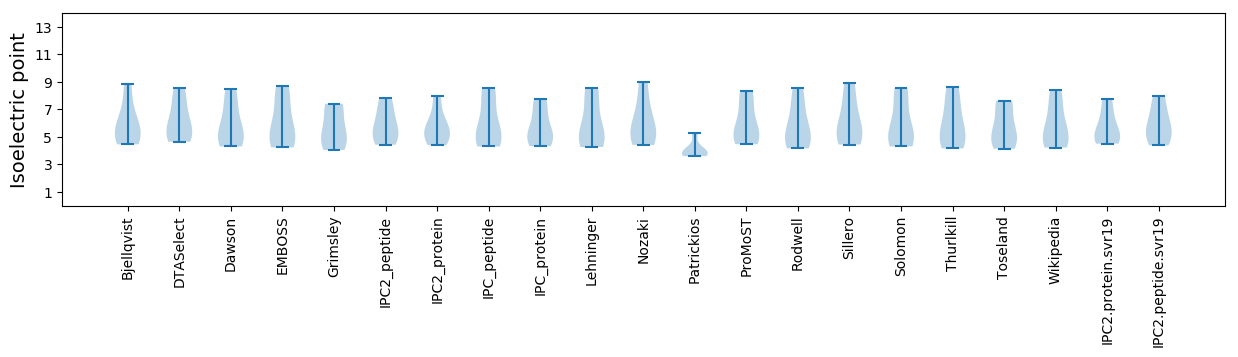
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W8L6|A0A4P8W8L6_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_445 OX=2585457 PE=4 SV=1
MM1 pKa = 7.51KK2 pKa = 9.74PCLHH6 pKa = 7.04PSLVVVLGDD15 pKa = 4.54NPDD18 pKa = 3.47TGKK21 pKa = 10.86KK22 pKa = 9.47IIRR25 pKa = 11.84FRR27 pKa = 11.84GRR29 pKa = 11.84SEE31 pKa = 4.04DD32 pKa = 4.08TIQSLSDD39 pKa = 3.16LYY41 pKa = 11.38GRR43 pKa = 11.84EE44 pKa = 3.98NVLLIPCGQCEE55 pKa = 4.28SCRR58 pKa = 11.84INYY61 pKa = 8.16AQVWSIRR68 pKa = 11.84CSLEE72 pKa = 4.45AMMTPYY78 pKa = 10.38HH79 pKa = 5.96YY80 pKa = 10.52FITLTYY86 pKa = 10.87DD87 pKa = 3.38DD88 pKa = 4.32LHH90 pKa = 8.08YY91 pKa = 10.94PFANLSDD98 pKa = 3.5FTKK101 pKa = 9.63WMKK104 pKa = 10.51RR105 pKa = 11.84MRR107 pKa = 11.84KK108 pKa = 9.28NYY110 pKa = 8.51PDD112 pKa = 3.22HH113 pKa = 6.3HH114 pKa = 6.67FKK116 pKa = 11.29YY117 pKa = 10.15FMSMEE122 pKa = 4.11RR123 pKa = 11.84GEE125 pKa = 4.37RR126 pKa = 11.84TNRR129 pKa = 11.84LHH131 pKa = 5.94FHH133 pKa = 6.79VILFSQIALSLSDD146 pKa = 3.46GKK148 pKa = 10.83FINGFMRR155 pKa = 11.84YY156 pKa = 8.16KK157 pKa = 10.52AKK159 pKa = 10.43EE160 pKa = 3.87IQSTWNLGVNNEE172 pKa = 3.88VSEE175 pKa = 4.5FEE177 pKa = 4.43CACAKK182 pKa = 10.28YY183 pKa = 9.28VAKK186 pKa = 9.42YY187 pKa = 7.8TSKK190 pKa = 10.9NGLVRR195 pKa = 11.84MSRR198 pKa = 11.84NLGKK202 pKa = 10.49SYY204 pKa = 10.96VLSHH208 pKa = 6.73ASEE211 pKa = 3.95IVADD215 pKa = 3.86NFKK218 pKa = 11.09VYY220 pKa = 11.19GDD222 pKa = 3.56FSDD225 pKa = 4.56RR226 pKa = 11.84NRR228 pKa = 11.84YY229 pKa = 7.23WVNLPSQCALWLLDD243 pKa = 4.02FYY245 pKa = 11.25EE246 pKa = 5.12SNVLSYY252 pKa = 10.96KK253 pKa = 10.53EE254 pKa = 3.96SLQVLSHH261 pKa = 6.69LKK263 pKa = 10.16QIYY266 pKa = 9.66SMKK269 pKa = 10.08NRR271 pKa = 11.84CSTCEE276 pKa = 4.31DD277 pKa = 3.44IEE279 pKa = 4.69VIDD282 pKa = 4.47TIYY285 pKa = 10.93DD286 pKa = 3.54EE287 pKa = 4.5KK288 pKa = 11.12NKK290 pKa = 10.08KK291 pKa = 10.12RR292 pKa = 11.84GVKK295 pKa = 10.14GCLL298 pKa = 3.1
MM1 pKa = 7.51KK2 pKa = 9.74PCLHH6 pKa = 7.04PSLVVVLGDD15 pKa = 4.54NPDD18 pKa = 3.47TGKK21 pKa = 10.86KK22 pKa = 9.47IIRR25 pKa = 11.84FRR27 pKa = 11.84GRR29 pKa = 11.84SEE31 pKa = 4.04DD32 pKa = 4.08TIQSLSDD39 pKa = 3.16LYY41 pKa = 11.38GRR43 pKa = 11.84EE44 pKa = 3.98NVLLIPCGQCEE55 pKa = 4.28SCRR58 pKa = 11.84INYY61 pKa = 8.16AQVWSIRR68 pKa = 11.84CSLEE72 pKa = 4.45AMMTPYY78 pKa = 10.38HH79 pKa = 5.96YY80 pKa = 10.52FITLTYY86 pKa = 10.87DD87 pKa = 3.38DD88 pKa = 4.32LHH90 pKa = 8.08YY91 pKa = 10.94PFANLSDD98 pKa = 3.5FTKK101 pKa = 9.63WMKK104 pKa = 10.51RR105 pKa = 11.84MRR107 pKa = 11.84KK108 pKa = 9.28NYY110 pKa = 8.51PDD112 pKa = 3.22HH113 pKa = 6.3HH114 pKa = 6.67FKK116 pKa = 11.29YY117 pKa = 10.15FMSMEE122 pKa = 4.11RR123 pKa = 11.84GEE125 pKa = 4.37RR126 pKa = 11.84TNRR129 pKa = 11.84LHH131 pKa = 5.94FHH133 pKa = 6.79VILFSQIALSLSDD146 pKa = 3.46GKK148 pKa = 10.83FINGFMRR155 pKa = 11.84YY156 pKa = 8.16KK157 pKa = 10.52AKK159 pKa = 10.43EE160 pKa = 3.87IQSTWNLGVNNEE172 pKa = 3.88VSEE175 pKa = 4.5FEE177 pKa = 4.43CACAKK182 pKa = 10.28YY183 pKa = 9.28VAKK186 pKa = 9.42YY187 pKa = 7.8TSKK190 pKa = 10.9NGLVRR195 pKa = 11.84MSRR198 pKa = 11.84NLGKK202 pKa = 10.49SYY204 pKa = 10.96VLSHH208 pKa = 6.73ASEE211 pKa = 3.95IVADD215 pKa = 3.86NFKK218 pKa = 11.09VYY220 pKa = 11.19GDD222 pKa = 3.56FSDD225 pKa = 4.56RR226 pKa = 11.84NRR228 pKa = 11.84YY229 pKa = 7.23WVNLPSQCALWLLDD243 pKa = 4.02FYY245 pKa = 11.25EE246 pKa = 5.12SNVLSYY252 pKa = 10.96KK253 pKa = 10.53EE254 pKa = 3.96SLQVLSHH261 pKa = 6.69LKK263 pKa = 10.16QIYY266 pKa = 9.66SMKK269 pKa = 10.08NRR271 pKa = 11.84CSTCEE276 pKa = 4.31DD277 pKa = 3.44IEE279 pKa = 4.69VIDD282 pKa = 4.47TIYY285 pKa = 10.93DD286 pKa = 3.54EE287 pKa = 4.5KK288 pKa = 11.12NKK290 pKa = 10.08KK291 pKa = 10.12RR292 pKa = 11.84GVKK295 pKa = 10.14GCLL298 pKa = 3.1
Molecular weight: 34.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1378 |
88 |
559 |
229.7 |
25.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.894 ± 0.897 | 1.742 ± 0.578 |
6.531 ± 0.632 | 4.862 ± 0.411 |
5.007 ± 0.44 | 5.806 ± 1.015 |
1.742 ± 0.416 | 6.386 ± 1.296 |
6.313 ± 0.682 | 7.983 ± 0.727 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.83 ± 0.281 | 5.66 ± 0.752 |
3.846 ± 0.769 | 3.701 ± 0.373 |
3.846 ± 0.643 | 9.579 ± 0.761 |
5.443 ± 0.844 | 5.66 ± 0.701 |
1.451 ± 0.308 | 4.717 ± 0.759 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
