
Takifugu flavidus (sansaifugu)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei;
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
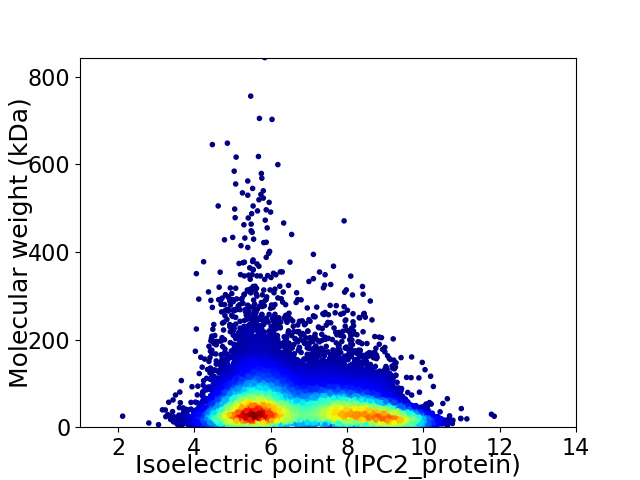
Virtual 2D-PAGE plot for 29076 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
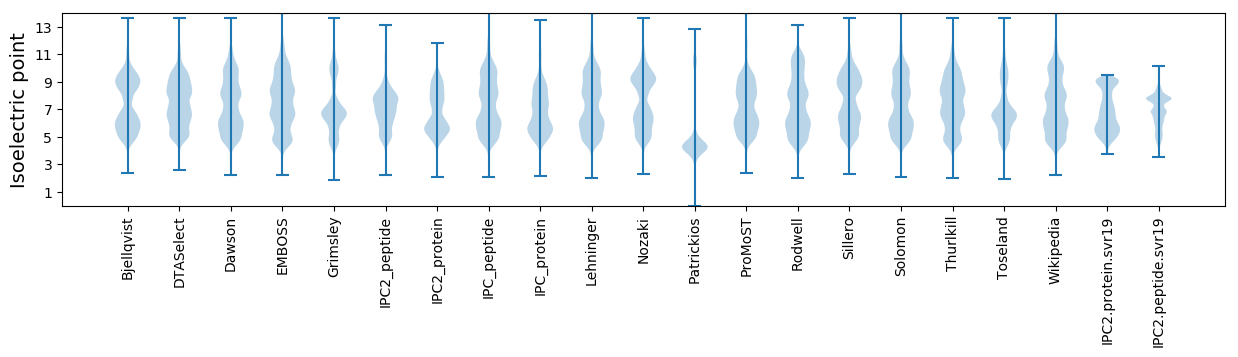
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C6NUM2|A0A5C6NUM2_9TELE Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6 OS=Takifugu flavidus OX=433684 GN=D4764_18G0013140 PE=4 SV=1
MM1 pKa = 6.6QQVEE5 pKa = 4.59EE6 pKa = 4.37AAMTTQPSPSEE17 pKa = 3.75QAPPLTEE24 pKa = 3.93SPEE27 pKa = 4.37AKK29 pKa = 10.18DD30 pKa = 3.43HH31 pKa = 6.24QQDD34 pKa = 3.75QNHH37 pKa = 5.87LTVISYY43 pKa = 10.76DD44 pKa = 3.92SEE46 pKa = 4.31PSGAGAPVQSTSPKK60 pKa = 7.82PAGRR64 pKa = 11.84SEE66 pKa = 3.62EE67 pKa = 4.17RR68 pKa = 11.84ANGRR72 pKa = 11.84PGLGSRR78 pKa = 11.84SSSVAGGSPRR88 pKa = 11.84PSLARR93 pKa = 11.84QPSTLTEE100 pKa = 3.9AALDD104 pKa = 3.9GSKK107 pKa = 10.64PRR109 pKa = 11.84DD110 pKa = 3.64YY111 pKa = 11.33LFLAILSCFCPLWPINIVALAFSVMSRR138 pKa = 11.84NSLQQGNVDD147 pKa = 3.32GARR150 pKa = 11.84RR151 pKa = 11.84LGRR154 pKa = 11.84NAMILSIVSILGGIAAIAATIALNWGPTDD183 pKa = 4.28APTGLCSTDD192 pKa = 3.58TSCLTIPWSQPSDD205 pKa = 3.76CPTDD209 pKa = 3.75PEE211 pKa = 4.51LTTDD215 pKa = 4.33AGITTDD221 pKa = 4.36PGLTTDD227 pKa = 5.04AGITTDD233 pKa = 4.36PGLTTDD239 pKa = 4.99AGLTTDD245 pKa = 4.96PEE247 pKa = 4.46LTTDD251 pKa = 4.57AEE253 pKa = 4.36LSGCFTDD260 pKa = 5.92PEE262 pKa = 4.54LTTDD266 pKa = 4.01AGVSGCFTDD275 pKa = 5.53PEE277 pKa = 4.52LTTDD281 pKa = 3.93PGLNTDD287 pKa = 4.42PEE289 pKa = 4.53LTTDD293 pKa = 4.28AGLTTDD299 pKa = 4.96PEE301 pKa = 4.46LTTDD305 pKa = 4.35AEE307 pKa = 4.49LTTEE311 pKa = 4.81AEE313 pKa = 4.17LSGCFTDD320 pKa = 5.92PEE322 pKa = 4.49LTTDD326 pKa = 4.57AEE328 pKa = 4.36LSGCFTDD335 pKa = 5.92PEE337 pKa = 4.54LTTDD341 pKa = 4.01AGVSGCFTDD350 pKa = 5.53PEE352 pKa = 4.54LTTDD356 pKa = 4.01AGVSGCFTDD365 pKa = 5.53PEE367 pKa = 4.54LTTDD371 pKa = 4.01AGVSGCFTDD380 pKa = 5.53PEE382 pKa = 4.49LTTDD386 pKa = 4.57AEE388 pKa = 4.36LSGCFTDD395 pKa = 5.92PEE397 pKa = 4.54LTTDD401 pKa = 4.01AGVSGCFTDD410 pKa = 4.54PEE412 pKa = 4.48LPKK415 pKa = 10.71LFSILPADD423 pKa = 3.67VTVCYY428 pKa = 10.32HH429 pKa = 6.95LLL431 pKa = 3.8
MM1 pKa = 6.6QQVEE5 pKa = 4.59EE6 pKa = 4.37AAMTTQPSPSEE17 pKa = 3.75QAPPLTEE24 pKa = 3.93SPEE27 pKa = 4.37AKK29 pKa = 10.18DD30 pKa = 3.43HH31 pKa = 6.24QQDD34 pKa = 3.75QNHH37 pKa = 5.87LTVISYY43 pKa = 10.76DD44 pKa = 3.92SEE46 pKa = 4.31PSGAGAPVQSTSPKK60 pKa = 7.82PAGRR64 pKa = 11.84SEE66 pKa = 3.62EE67 pKa = 4.17RR68 pKa = 11.84ANGRR72 pKa = 11.84PGLGSRR78 pKa = 11.84SSSVAGGSPRR88 pKa = 11.84PSLARR93 pKa = 11.84QPSTLTEE100 pKa = 3.9AALDD104 pKa = 3.9GSKK107 pKa = 10.64PRR109 pKa = 11.84DD110 pKa = 3.64YY111 pKa = 11.33LFLAILSCFCPLWPINIVALAFSVMSRR138 pKa = 11.84NSLQQGNVDD147 pKa = 3.32GARR150 pKa = 11.84RR151 pKa = 11.84LGRR154 pKa = 11.84NAMILSIVSILGGIAAIAATIALNWGPTDD183 pKa = 4.28APTGLCSTDD192 pKa = 3.58TSCLTIPWSQPSDD205 pKa = 3.76CPTDD209 pKa = 3.75PEE211 pKa = 4.51LTTDD215 pKa = 4.33AGITTDD221 pKa = 4.36PGLTTDD227 pKa = 5.04AGITTDD233 pKa = 4.36PGLTTDD239 pKa = 4.99AGLTTDD245 pKa = 4.96PEE247 pKa = 4.46LTTDD251 pKa = 4.57AEE253 pKa = 4.36LSGCFTDD260 pKa = 5.92PEE262 pKa = 4.54LTTDD266 pKa = 4.01AGVSGCFTDD275 pKa = 5.53PEE277 pKa = 4.52LTTDD281 pKa = 3.93PGLNTDD287 pKa = 4.42PEE289 pKa = 4.53LTTDD293 pKa = 4.28AGLTTDD299 pKa = 4.96PEE301 pKa = 4.46LTTDD305 pKa = 4.35AEE307 pKa = 4.49LTTEE311 pKa = 4.81AEE313 pKa = 4.17LSGCFTDD320 pKa = 5.92PEE322 pKa = 4.49LTTDD326 pKa = 4.57AEE328 pKa = 4.36LSGCFTDD335 pKa = 5.92PEE337 pKa = 4.54LTTDD341 pKa = 4.01AGVSGCFTDD350 pKa = 5.53PEE352 pKa = 4.54LTTDD356 pKa = 4.01AGVSGCFTDD365 pKa = 5.53PEE367 pKa = 4.54LTTDD371 pKa = 4.01AGVSGCFTDD380 pKa = 5.53PEE382 pKa = 4.49LTTDD386 pKa = 4.57AEE388 pKa = 4.36LSGCFTDD395 pKa = 5.92PEE397 pKa = 4.54LTTDD401 pKa = 4.01AGVSGCFTDD410 pKa = 4.54PEE412 pKa = 4.48LPKK415 pKa = 10.71LFSILPADD423 pKa = 3.67VTVCYY428 pKa = 10.32HH429 pKa = 6.95LLL431 pKa = 3.8
Molecular weight: 44.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C6MT66|A0A5C6MT66_9TELE Major facilitator superfamily domain-containing protein 5 OS=Takifugu flavidus OX=433684 GN=D4764_08G0006570 PE=3 SV=1
MM1 pKa = 7.0PAKK4 pKa = 10.27RR5 pKa = 11.84RR6 pKa = 11.84ISKK9 pKa = 10.66AKK11 pKa = 8.41TSTVRR16 pKa = 11.84EE17 pKa = 4.24RR18 pKa = 11.84ILQAVSNSRR27 pKa = 11.84EE28 pKa = 4.06RR29 pKa = 11.84NGVPLSTLKK38 pKa = 10.67KK39 pKa = 8.97ALAAGGYY46 pKa = 10.31DD47 pKa = 3.42VVKK50 pKa = 10.8NKK52 pKa = 10.58ASVKK56 pKa = 9.45IAIRR60 pKa = 11.84NLLNRR65 pKa = 11.84GALVQTRR72 pKa = 11.84GFGASAAFKK81 pKa = 10.31INKK84 pKa = 8.54NVAGAIAKK92 pKa = 10.02RR93 pKa = 11.84PRR95 pKa = 11.84GKK97 pKa = 10.14KK98 pKa = 9.04AGRR101 pKa = 11.84RR102 pKa = 11.84GRR104 pKa = 11.84KK105 pKa = 8.35RR106 pKa = 11.84AARR109 pKa = 11.84RR110 pKa = 11.84PGKK113 pKa = 9.99KK114 pKa = 9.51GRR116 pKa = 11.84ARR118 pKa = 11.84KK119 pKa = 9.14RR120 pKa = 11.84AGAKK124 pKa = 9.71KK125 pKa = 9.4SRR127 pKa = 11.84KK128 pKa = 8.3RR129 pKa = 11.84GKK131 pKa = 9.99RR132 pKa = 11.84SRR134 pKa = 11.84KK135 pKa = 9.05RR136 pKa = 11.84AAKK139 pKa = 9.83RR140 pKa = 11.84PKK142 pKa = 10.01RR143 pKa = 11.84SAKK146 pKa = 8.77GRR148 pKa = 11.84KK149 pKa = 8.92RR150 pKa = 11.84RR151 pKa = 11.84GKK153 pKa = 9.66KK154 pKa = 9.6AAGKK158 pKa = 9.77RR159 pKa = 11.84KK160 pKa = 9.21SRR162 pKa = 11.84KK163 pKa = 9.35AGTKK167 pKa = 8.8RR168 pKa = 11.84RR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 3.55
MM1 pKa = 7.0PAKK4 pKa = 10.27RR5 pKa = 11.84RR6 pKa = 11.84ISKK9 pKa = 10.66AKK11 pKa = 8.41TSTVRR16 pKa = 11.84EE17 pKa = 4.24RR18 pKa = 11.84ILQAVSNSRR27 pKa = 11.84EE28 pKa = 4.06RR29 pKa = 11.84NGVPLSTLKK38 pKa = 10.67KK39 pKa = 8.97ALAAGGYY46 pKa = 10.31DD47 pKa = 3.42VVKK50 pKa = 10.8NKK52 pKa = 10.58ASVKK56 pKa = 9.45IAIRR60 pKa = 11.84NLLNRR65 pKa = 11.84GALVQTRR72 pKa = 11.84GFGASAAFKK81 pKa = 10.31INKK84 pKa = 8.54NVAGAIAKK92 pKa = 10.02RR93 pKa = 11.84PRR95 pKa = 11.84GKK97 pKa = 10.14KK98 pKa = 9.04AGRR101 pKa = 11.84RR102 pKa = 11.84GRR104 pKa = 11.84KK105 pKa = 8.35RR106 pKa = 11.84AARR109 pKa = 11.84RR110 pKa = 11.84PGKK113 pKa = 9.99KK114 pKa = 9.51GRR116 pKa = 11.84ARR118 pKa = 11.84KK119 pKa = 9.14RR120 pKa = 11.84AGAKK124 pKa = 9.71KK125 pKa = 9.4SRR127 pKa = 11.84KK128 pKa = 8.3RR129 pKa = 11.84GKK131 pKa = 9.99RR132 pKa = 11.84SRR134 pKa = 11.84KK135 pKa = 9.05RR136 pKa = 11.84AAKK139 pKa = 9.83RR140 pKa = 11.84PKK142 pKa = 10.01RR143 pKa = 11.84SAKK146 pKa = 8.77GRR148 pKa = 11.84KK149 pKa = 8.92RR150 pKa = 11.84RR151 pKa = 11.84GKK153 pKa = 9.66KK154 pKa = 9.6AAGKK158 pKa = 9.77RR159 pKa = 11.84KK160 pKa = 9.21SRR162 pKa = 11.84KK163 pKa = 9.35AGTKK167 pKa = 8.8RR168 pKa = 11.84RR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 3.55
Molecular weight: 18.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14023189 |
8 |
23532 |
482.3 |
53.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.684 ± 0.014 | 2.273 ± 0.011 |
5.044 ± 0.011 | 6.853 ± 0.02 |
3.473 ± 0.01 | 6.506 ± 0.02 |
2.73 ± 0.011 | 4.157 ± 0.011 |
5.262 ± 0.016 | 9.772 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.293 ± 0.007 | 3.598 ± 0.012 |
5.91 ± 0.02 | 4.69 ± 0.016 |
6.211 ± 0.016 | 8.824 ± 0.019 |
5.529 ± 0.013 | 6.44 ± 0.014 |
1.241 ± 0.005 | 2.51 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
