
Lucerne transient streak virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Sobemovirus
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
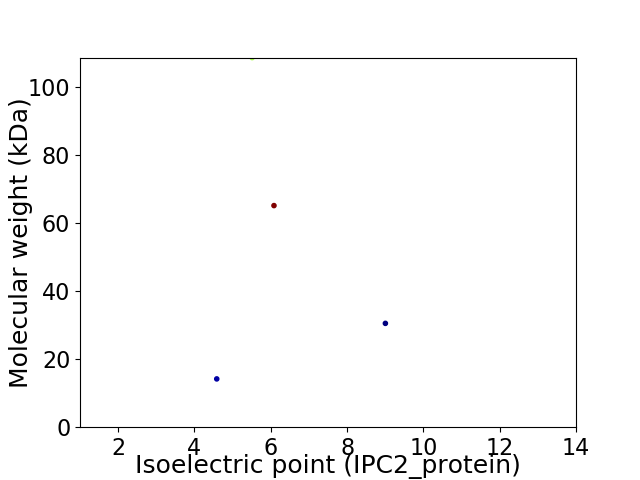
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
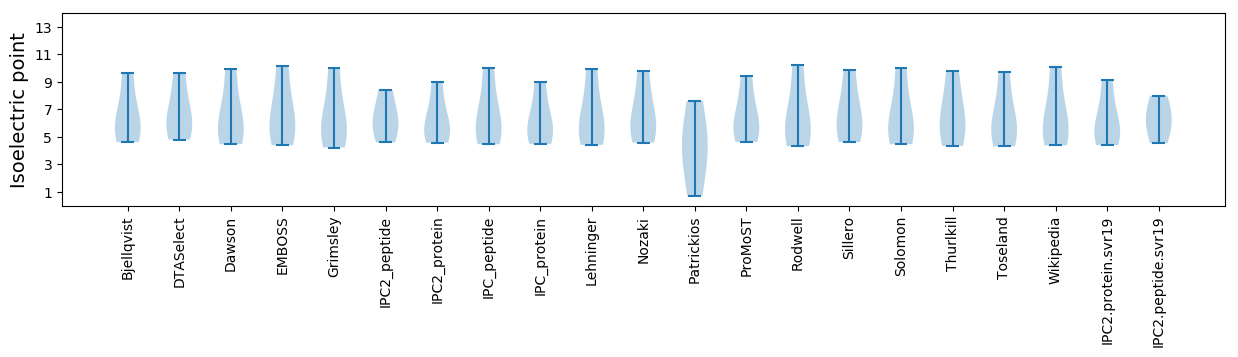
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L0MN56|L0MN56_9VIRU N-terminal protein OS=Lucerne transient streak virus OX=12470 PE=4 SV=1
MM1 pKa = 7.64PSVIVEE7 pKa = 4.51CYY9 pKa = 7.1TTDD12 pKa = 2.9SHH14 pKa = 8.52RR15 pKa = 11.84DD16 pKa = 3.11IPLFSQKK23 pKa = 8.69IWFDD27 pKa = 3.51NVDD30 pKa = 4.34EE31 pKa = 4.38LTSDD35 pKa = 3.5YY36 pKa = 10.96VRR38 pKa = 11.84VYY40 pKa = 10.49RR41 pKa = 11.84AVRR44 pKa = 11.84DD45 pKa = 3.79YY46 pKa = 11.87SKK48 pKa = 11.12VDD50 pKa = 3.64YY51 pKa = 11.07IEE53 pKa = 4.62VEE55 pKa = 4.07LRR57 pKa = 11.84CNFCHH62 pKa = 6.66YY63 pKa = 10.32YY64 pKa = 10.09NSRR67 pKa = 11.84GQIVQRR73 pKa = 11.84YY74 pKa = 8.16LGCKK78 pKa = 9.69DD79 pKa = 3.26VEE81 pKa = 4.17IRR83 pKa = 11.84NISLPDD89 pKa = 3.68NDD91 pKa = 4.64PYY93 pKa = 11.24TYY95 pKa = 9.83TVYY98 pKa = 10.88SVDD101 pKa = 3.52CAICRR106 pKa = 11.84KK107 pKa = 9.79IPTEE111 pKa = 4.23SEE113 pKa = 4.32DD114 pKa = 3.8EE115 pKa = 4.27SDD117 pKa = 4.24SSFF120 pKa = 3.63
MM1 pKa = 7.64PSVIVEE7 pKa = 4.51CYY9 pKa = 7.1TTDD12 pKa = 2.9SHH14 pKa = 8.52RR15 pKa = 11.84DD16 pKa = 3.11IPLFSQKK23 pKa = 8.69IWFDD27 pKa = 3.51NVDD30 pKa = 4.34EE31 pKa = 4.38LTSDD35 pKa = 3.5YY36 pKa = 10.96VRR38 pKa = 11.84VYY40 pKa = 10.49RR41 pKa = 11.84AVRR44 pKa = 11.84DD45 pKa = 3.79YY46 pKa = 11.87SKK48 pKa = 11.12VDD50 pKa = 3.64YY51 pKa = 11.07IEE53 pKa = 4.62VEE55 pKa = 4.07LRR57 pKa = 11.84CNFCHH62 pKa = 6.66YY63 pKa = 10.32YY64 pKa = 10.09NSRR67 pKa = 11.84GQIVQRR73 pKa = 11.84YY74 pKa = 8.16LGCKK78 pKa = 9.69DD79 pKa = 3.26VEE81 pKa = 4.17IRR83 pKa = 11.84NISLPDD89 pKa = 3.68NDD91 pKa = 4.64PYY93 pKa = 11.24TYY95 pKa = 9.83TVYY98 pKa = 10.88SVDD101 pKa = 3.52CAICRR106 pKa = 11.84KK107 pKa = 9.79IPTEE111 pKa = 4.23SEE113 pKa = 4.32DD114 pKa = 3.8EE115 pKa = 4.27SDD117 pKa = 4.24SSFF120 pKa = 3.63
Molecular weight: 14.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L0MPW1|L0MPW1_9VIRU Capsid protein OS=Lucerne transient streak virus OX=12470 PE=3 SV=1
MM1 pKa = 6.65VAKK4 pKa = 9.86TRR6 pKa = 11.84RR7 pKa = 11.84ANRR10 pKa = 11.84KK11 pKa = 8.93KK12 pKa = 10.4KK13 pKa = 10.44RR14 pKa = 11.84NNNTQRR20 pKa = 11.84RR21 pKa = 11.84TTEE24 pKa = 3.88PNRR27 pKa = 11.84QLATARR33 pKa = 11.84RR34 pKa = 11.84VTIPAATSVVVRR46 pKa = 11.84KK47 pKa = 9.61SSPVIRR53 pKa = 11.84SDD55 pKa = 2.91RR56 pKa = 11.84KK57 pKa = 10.45GIQVCNTEE65 pKa = 5.1FIRR68 pKa = 11.84SFDD71 pKa = 3.88LNAGFTVGSLPCTPGTMPWLSRR93 pKa = 11.84MAASWGRR100 pKa = 11.84WKK102 pKa = 10.37WNSLRR107 pKa = 11.84FTYY110 pKa = 10.26IPAAPSSTQGTVAMGFLYY128 pKa = 10.6DD129 pKa = 4.97ALDD132 pKa = 3.93SLPSNLASMSSLDD145 pKa = 3.59GFTTGAVWSGCEE157 pKa = 3.75GSAYY161 pKa = 10.07LAKK164 pKa = 10.32PSMRR168 pKa = 11.84DD169 pKa = 2.9IPGAISTCLDD179 pKa = 3.46IDD181 pKa = 4.02PDD183 pKa = 3.41WRR185 pKa = 11.84QKK187 pKa = 7.83RR188 pKa = 11.84WRR190 pKa = 11.84YY191 pKa = 7.65ITALEE196 pKa = 4.01LAALSKK202 pKa = 10.83PDD204 pKa = 3.77FNIYY208 pKa = 10.54APARR212 pKa = 11.84LAIAAANSSINIASVGTLYY231 pKa = 9.99ATYY234 pKa = 9.79EE235 pKa = 4.1VEE237 pKa = 4.82LFDD240 pKa = 6.38PISAEE245 pKa = 4.17LNLPTPAVVTQAKK258 pKa = 10.15EE259 pKa = 3.9DD260 pKa = 4.06TTKK263 pKa = 9.41HH264 pKa = 4.8TSNTEE269 pKa = 3.94EE270 pKa = 4.25EE271 pKa = 4.27DD272 pKa = 3.89GKK274 pKa = 11.08AKK276 pKa = 10.23PHH278 pKa = 6.01
MM1 pKa = 6.65VAKK4 pKa = 9.86TRR6 pKa = 11.84RR7 pKa = 11.84ANRR10 pKa = 11.84KK11 pKa = 8.93KK12 pKa = 10.4KK13 pKa = 10.44RR14 pKa = 11.84NNNTQRR20 pKa = 11.84RR21 pKa = 11.84TTEE24 pKa = 3.88PNRR27 pKa = 11.84QLATARR33 pKa = 11.84RR34 pKa = 11.84VTIPAATSVVVRR46 pKa = 11.84KK47 pKa = 9.61SSPVIRR53 pKa = 11.84SDD55 pKa = 2.91RR56 pKa = 11.84KK57 pKa = 10.45GIQVCNTEE65 pKa = 5.1FIRR68 pKa = 11.84SFDD71 pKa = 3.88LNAGFTVGSLPCTPGTMPWLSRR93 pKa = 11.84MAASWGRR100 pKa = 11.84WKK102 pKa = 10.37WNSLRR107 pKa = 11.84FTYY110 pKa = 10.26IPAAPSSTQGTVAMGFLYY128 pKa = 10.6DD129 pKa = 4.97ALDD132 pKa = 3.93SLPSNLASMSSLDD145 pKa = 3.59GFTTGAVWSGCEE157 pKa = 3.75GSAYY161 pKa = 10.07LAKK164 pKa = 10.32PSMRR168 pKa = 11.84DD169 pKa = 2.9IPGAISTCLDD179 pKa = 3.46IDD181 pKa = 4.02PDD183 pKa = 3.41WRR185 pKa = 11.84QKK187 pKa = 7.83RR188 pKa = 11.84WRR190 pKa = 11.84YY191 pKa = 7.65ITALEE196 pKa = 4.01LAALSKK202 pKa = 10.83PDD204 pKa = 3.77FNIYY208 pKa = 10.54APARR212 pKa = 11.84LAIAAANSSINIASVGTLYY231 pKa = 9.99ATYY234 pKa = 9.79EE235 pKa = 4.1VEE237 pKa = 4.82LFDD240 pKa = 6.38PISAEE245 pKa = 4.17LNLPTPAVVTQAKK258 pKa = 10.15EE259 pKa = 3.9DD260 pKa = 4.06TTKK263 pKa = 9.41HH264 pKa = 4.8TSNTEE269 pKa = 3.94EE270 pKa = 4.25EE271 pKa = 4.27DD272 pKa = 3.89GKK274 pKa = 11.08AKK276 pKa = 10.23PHH278 pKa = 6.01
Molecular weight: 30.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1983 |
120 |
982 |
495.8 |
54.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.413 ± 1.046 | 2.118 ± 0.337 |
5.093 ± 0.535 | 7.211 ± 0.991 |
2.471 ± 0.316 | 7.665 ± 1.002 |
1.261 ± 0.184 | 4.488 ± 0.453 |
6.001 ± 0.504 | 8.926 ± 0.734 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.614 ± 0.118 | 3.48 ± 0.39 |
5.547 ± 0.288 | 3.076 ± 0.319 |
5.547 ± 0.704 | 9.934 ± 1.093 |
5.497 ± 1.048 | 7.211 ± 0.503 |
2.118 ± 0.135 | 3.328 ± 0.533 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
