
Pepper cryptic virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Deltapartitivirus
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
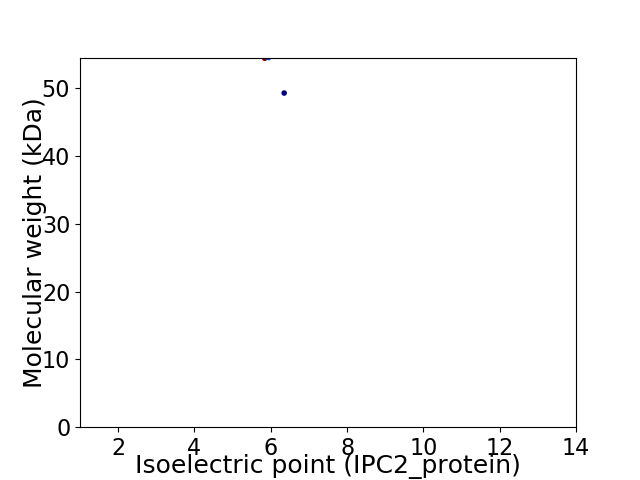
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1J1DT58|A0A1J1DT58_9VIRU Putative RNA-dependent RNA polymerase OS=Pepper cryptic virus 2 OX=1050903 GN=RdRp PE=4 SV=1
MM1 pKa = 7.79AIYY4 pKa = 10.79RR5 pKa = 11.84MLNGYY10 pKa = 10.41AFTAFGNDD18 pKa = 4.26LEE20 pKa = 5.35KK21 pKa = 10.73LDD23 pKa = 3.66QRR25 pKa = 11.84HH26 pKa = 4.27IHH28 pKa = 6.55RR29 pKa = 11.84IRR31 pKa = 11.84RR32 pKa = 11.84EE33 pKa = 3.54EE34 pKa = 3.67ATTYY38 pKa = 10.64RR39 pKa = 11.84DD40 pKa = 3.18EE41 pKa = 4.53FALKK45 pKa = 10.55EE46 pKa = 3.98LMDD49 pKa = 5.19LNPILYY55 pKa = 9.57EE56 pKa = 3.8QFLEE60 pKa = 4.64GWSRR64 pKa = 11.84SYY66 pKa = 11.77YY67 pKa = 10.24EE68 pKa = 4.29GSKK71 pKa = 10.22HH72 pKa = 5.43LQAIIQYY79 pKa = 7.92GIPDD83 pKa = 4.1PHH85 pKa = 7.13PNLIDD90 pKa = 3.52KK91 pKa = 10.02EE92 pKa = 4.39IYY94 pKa = 9.89NKK96 pKa = 10.4AGYY99 pKa = 9.56VVLEE103 pKa = 4.36SLGSLPRR110 pKa = 11.84VRR112 pKa = 11.84AFDD115 pKa = 3.53VLTEE119 pKa = 4.16LDD121 pKa = 3.86SVHH124 pKa = 6.32YY125 pKa = 8.51EE126 pKa = 3.88QSSSAGCDD134 pKa = 3.13YY135 pKa = 10.74HH136 pKa = 8.59GPKK139 pKa = 10.71GPIQGEE145 pKa = 3.89NHH147 pKa = 5.33IRR149 pKa = 11.84AITRR153 pKa = 11.84AKK155 pKa = 8.72ATLWSAIKK163 pKa = 10.61DD164 pKa = 3.37EE165 pKa = 4.83GEE167 pKa = 4.44GIEE170 pKa = 4.1HH171 pKa = 7.42VIRR174 pKa = 11.84SSVPDD179 pKa = 3.2VGYY182 pKa = 8.77TRR184 pKa = 11.84TQLTDD189 pKa = 3.66LYY191 pKa = 9.69EE192 pKa = 3.99KK193 pKa = 10.34TKK195 pKa = 10.51IRR197 pKa = 11.84GVWGRR202 pKa = 11.84AFHH205 pKa = 6.67YY206 pKa = 10.13ILLEE210 pKa = 4.04GTVANPLLDD219 pKa = 3.47VFKK222 pKa = 10.96RR223 pKa = 11.84GGTFYY228 pKa = 10.77HH229 pKa = 7.19IGEE232 pKa = 4.3NPQYY236 pKa = 10.44SVPDD240 pKa = 3.41ILSQVSEE247 pKa = 4.21CCKK250 pKa = 10.48YY251 pKa = 10.65LVAIDD256 pKa = 4.01WSNFDD261 pKa = 3.1ATVARR266 pKa = 11.84FEE268 pKa = 4.61INMAFDD274 pKa = 5.15LIKK277 pKa = 10.0TLIMFPNIEE286 pKa = 4.17TEE288 pKa = 4.16LCFEE292 pKa = 4.47ICRR295 pKa = 11.84QLFIHH300 pKa = 6.58KK301 pKa = 9.99KK302 pKa = 8.94IAAPDD307 pKa = 3.67GNIYY311 pKa = 9.66WSHH314 pKa = 7.32KK315 pKa = 9.34GIPSGSYY322 pKa = 7.09FTSIIGSIVNRR333 pKa = 11.84LRR335 pKa = 11.84VEE337 pKa = 4.08YY338 pKa = 9.96IFRR341 pKa = 11.84KK342 pKa = 10.15AYY344 pKa = 9.79GVGPKK349 pKa = 8.76MCYY352 pKa = 8.05TQGDD356 pKa = 4.0DD357 pKa = 3.95SLIGADD363 pKa = 4.25FRR365 pKa = 11.84VDD367 pKa = 3.45PDD369 pKa = 3.58RR370 pKa = 11.84LSEE373 pKa = 3.97IAAPLDD379 pKa = 3.48WKK381 pKa = 10.96LNPAKK386 pKa = 9.75TDD388 pKa = 3.14VSLYY392 pKa = 9.49PEE394 pKa = 4.16NVTFLGRR401 pKa = 11.84TMYY404 pKa = 10.87GGINQRR410 pKa = 11.84DD411 pKa = 4.03LKK413 pKa = 10.81RR414 pKa = 11.84CLRR417 pKa = 11.84LLIFPEE423 pKa = 4.57FPVPSGEE430 pKa = 3.56ISAYY434 pKa = 9.3RR435 pKa = 11.84AVSIAQDD442 pKa = 3.1AGGTSEE448 pKa = 4.4VLNSIAKK455 pKa = 9.81RR456 pKa = 11.84LRR458 pKa = 11.84RR459 pKa = 11.84QYY461 pKa = 10.98GVAEE465 pKa = 4.07EE466 pKa = 4.32HH467 pKa = 6.0TVPKK471 pKa = 10.45HH472 pKa = 5.48FEE474 pKa = 4.12LYY476 pKa = 10.74VPP478 pKa = 4.68
MM1 pKa = 7.79AIYY4 pKa = 10.79RR5 pKa = 11.84MLNGYY10 pKa = 10.41AFTAFGNDD18 pKa = 4.26LEE20 pKa = 5.35KK21 pKa = 10.73LDD23 pKa = 3.66QRR25 pKa = 11.84HH26 pKa = 4.27IHH28 pKa = 6.55RR29 pKa = 11.84IRR31 pKa = 11.84RR32 pKa = 11.84EE33 pKa = 3.54EE34 pKa = 3.67ATTYY38 pKa = 10.64RR39 pKa = 11.84DD40 pKa = 3.18EE41 pKa = 4.53FALKK45 pKa = 10.55EE46 pKa = 3.98LMDD49 pKa = 5.19LNPILYY55 pKa = 9.57EE56 pKa = 3.8QFLEE60 pKa = 4.64GWSRR64 pKa = 11.84SYY66 pKa = 11.77YY67 pKa = 10.24EE68 pKa = 4.29GSKK71 pKa = 10.22HH72 pKa = 5.43LQAIIQYY79 pKa = 7.92GIPDD83 pKa = 4.1PHH85 pKa = 7.13PNLIDD90 pKa = 3.52KK91 pKa = 10.02EE92 pKa = 4.39IYY94 pKa = 9.89NKK96 pKa = 10.4AGYY99 pKa = 9.56VVLEE103 pKa = 4.36SLGSLPRR110 pKa = 11.84VRR112 pKa = 11.84AFDD115 pKa = 3.53VLTEE119 pKa = 4.16LDD121 pKa = 3.86SVHH124 pKa = 6.32YY125 pKa = 8.51EE126 pKa = 3.88QSSSAGCDD134 pKa = 3.13YY135 pKa = 10.74HH136 pKa = 8.59GPKK139 pKa = 10.71GPIQGEE145 pKa = 3.89NHH147 pKa = 5.33IRR149 pKa = 11.84AITRR153 pKa = 11.84AKK155 pKa = 8.72ATLWSAIKK163 pKa = 10.61DD164 pKa = 3.37EE165 pKa = 4.83GEE167 pKa = 4.44GIEE170 pKa = 4.1HH171 pKa = 7.42VIRR174 pKa = 11.84SSVPDD179 pKa = 3.2VGYY182 pKa = 8.77TRR184 pKa = 11.84TQLTDD189 pKa = 3.66LYY191 pKa = 9.69EE192 pKa = 3.99KK193 pKa = 10.34TKK195 pKa = 10.51IRR197 pKa = 11.84GVWGRR202 pKa = 11.84AFHH205 pKa = 6.67YY206 pKa = 10.13ILLEE210 pKa = 4.04GTVANPLLDD219 pKa = 3.47VFKK222 pKa = 10.96RR223 pKa = 11.84GGTFYY228 pKa = 10.77HH229 pKa = 7.19IGEE232 pKa = 4.3NPQYY236 pKa = 10.44SVPDD240 pKa = 3.41ILSQVSEE247 pKa = 4.21CCKK250 pKa = 10.48YY251 pKa = 10.65LVAIDD256 pKa = 4.01WSNFDD261 pKa = 3.1ATVARR266 pKa = 11.84FEE268 pKa = 4.61INMAFDD274 pKa = 5.15LIKK277 pKa = 10.0TLIMFPNIEE286 pKa = 4.17TEE288 pKa = 4.16LCFEE292 pKa = 4.47ICRR295 pKa = 11.84QLFIHH300 pKa = 6.58KK301 pKa = 9.99KK302 pKa = 8.94IAAPDD307 pKa = 3.67GNIYY311 pKa = 9.66WSHH314 pKa = 7.32KK315 pKa = 9.34GIPSGSYY322 pKa = 7.09FTSIIGSIVNRR333 pKa = 11.84LRR335 pKa = 11.84VEE337 pKa = 4.08YY338 pKa = 9.96IFRR341 pKa = 11.84KK342 pKa = 10.15AYY344 pKa = 9.79GVGPKK349 pKa = 8.76MCYY352 pKa = 8.05TQGDD356 pKa = 4.0DD357 pKa = 3.95SLIGADD363 pKa = 4.25FRR365 pKa = 11.84VDD367 pKa = 3.45PDD369 pKa = 3.58RR370 pKa = 11.84LSEE373 pKa = 3.97IAAPLDD379 pKa = 3.48WKK381 pKa = 10.96LNPAKK386 pKa = 9.75TDD388 pKa = 3.14VSLYY392 pKa = 9.49PEE394 pKa = 4.16NVTFLGRR401 pKa = 11.84TMYY404 pKa = 10.87GGINQRR410 pKa = 11.84DD411 pKa = 4.03LKK413 pKa = 10.81RR414 pKa = 11.84CLRR417 pKa = 11.84LLIFPEE423 pKa = 4.57FPVPSGEE430 pKa = 3.56ISAYY434 pKa = 9.3RR435 pKa = 11.84AVSIAQDD442 pKa = 3.1AGGTSEE448 pKa = 4.4VLNSIAKK455 pKa = 9.81RR456 pKa = 11.84LRR458 pKa = 11.84RR459 pKa = 11.84QYY461 pKa = 10.98GVAEE465 pKa = 4.07EE466 pKa = 4.32HH467 pKa = 6.0TVPKK471 pKa = 10.45HH472 pKa = 5.48FEE474 pKa = 4.12LYY476 pKa = 10.74VPP478 pKa = 4.68
Molecular weight: 54.39 kDa
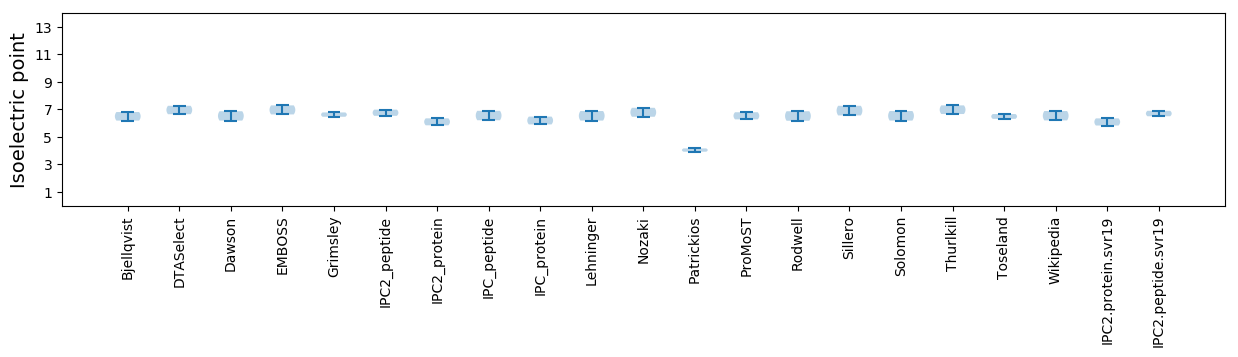
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1J1DT58|A0A1J1DT58_9VIRU Putative RNA-dependent RNA polymerase OS=Pepper cryptic virus 2 OX=1050903 GN=RdRp PE=4 SV=1
MM1 pKa = 7.31ATPVSDD7 pKa = 3.69TPTTKK12 pKa = 9.45STQDD16 pKa = 3.13KK17 pKa = 10.28TDD19 pKa = 3.5QLPAATPPITQKK31 pKa = 10.33PPSYY35 pKa = 9.81KK36 pKa = 10.28SEE38 pKa = 3.64ISRR41 pKa = 11.84IADD44 pKa = 3.54YY45 pKa = 10.96LADD48 pKa = 4.84DD49 pKa = 4.12YY50 pKa = 11.66EE51 pKa = 4.96IGYY54 pKa = 9.46VRR56 pKa = 11.84DD57 pKa = 3.71RR58 pKa = 11.84PPKK61 pKa = 7.61HH62 pKa = 6.13RR63 pKa = 11.84RR64 pKa = 11.84YY65 pKa = 10.28AVLRR69 pKa = 11.84TQALFNKK76 pKa = 10.12LIEE79 pKa = 4.47IYY81 pKa = 10.89GKK83 pKa = 10.22LFKK86 pKa = 10.93QNWEE90 pKa = 3.91FLRR93 pKa = 11.84RR94 pKa = 11.84SIDD97 pKa = 3.26GFTTAVGIRR106 pKa = 11.84EE107 pKa = 4.24RR108 pKa = 11.84QWEE111 pKa = 4.18NAARR115 pKa = 11.84AYY117 pKa = 9.43ISCWFFDD124 pKa = 3.95LYY126 pKa = 11.3VSVRR130 pKa = 11.84NAVEE134 pKa = 4.35KK135 pKa = 11.01VSGTAFTQYY144 pKa = 9.92YY145 pKa = 8.29HH146 pKa = 6.59AHH148 pKa = 6.6AAHH151 pKa = 7.12ILHH154 pKa = 7.08EE155 pKa = 4.27YY156 pKa = 10.23DD157 pKa = 4.0ALLVHH162 pKa = 6.82LNAIIRR168 pKa = 11.84PTHH171 pKa = 6.06IKK173 pKa = 10.3LALEE177 pKa = 3.99DD178 pKa = 4.55CIYY181 pKa = 10.29IPRR184 pKa = 11.84IATNMNITNANPFGLAEE201 pKa = 4.26ATWDD205 pKa = 3.51LDD207 pKa = 3.48ADD209 pKa = 4.36FAYY212 pKa = 11.02SIMDD216 pKa = 3.41RR217 pKa = 11.84MKK219 pKa = 10.88ASGSRR224 pKa = 11.84WRR226 pKa = 11.84VEE228 pKa = 3.77PLVTDD233 pKa = 3.69TLGRR237 pKa = 11.84PGWLFDD243 pKa = 3.24WHH245 pKa = 6.22QVGAQQQAYY254 pKa = 9.48AWFPSEE260 pKa = 4.46GNYY263 pKa = 11.0SMEE266 pKa = 4.8DD267 pKa = 3.88LIAPHH272 pKa = 7.14IIGEE276 pKa = 4.25ALTPLMGPCDD286 pKa = 3.9IDD288 pKa = 3.18DD289 pKa = 3.49WQYY292 pKa = 10.44FPNFIRR298 pKa = 11.84PANVPWQNMEE308 pKa = 3.96RR309 pKa = 11.84VSARR313 pKa = 11.84RR314 pKa = 11.84FRR316 pKa = 11.84GSAEE320 pKa = 3.47VRR322 pKa = 11.84TYY324 pKa = 11.4AEE326 pKa = 4.04EE327 pKa = 4.17KK328 pKa = 10.54VNLTRR333 pKa = 11.84FASTGHH339 pKa = 5.23EE340 pKa = 4.11TSKK343 pKa = 10.67KK344 pKa = 8.96RR345 pKa = 11.84SKK347 pKa = 10.47PSSTTTTSSQGGSRR361 pKa = 11.84DD362 pKa = 3.34MEE364 pKa = 4.46TEE366 pKa = 3.94DD367 pKa = 3.67EE368 pKa = 4.2PRR370 pKa = 11.84EE371 pKa = 4.1VVTDD375 pKa = 3.55ITLSQVTSTQQLQTLMTTGEE395 pKa = 3.97ADD397 pKa = 3.54RR398 pKa = 11.84FRR400 pKa = 11.84IIDD403 pKa = 3.52YY404 pKa = 8.86TYY406 pKa = 8.07YY407 pKa = 10.73CCVILRR413 pKa = 11.84NTAQKK418 pKa = 10.21RR419 pKa = 11.84DD420 pKa = 3.13GAIRR424 pKa = 11.84SFCFKK429 pKa = 10.54QQ430 pKa = 3.07
MM1 pKa = 7.31ATPVSDD7 pKa = 3.69TPTTKK12 pKa = 9.45STQDD16 pKa = 3.13KK17 pKa = 10.28TDD19 pKa = 3.5QLPAATPPITQKK31 pKa = 10.33PPSYY35 pKa = 9.81KK36 pKa = 10.28SEE38 pKa = 3.64ISRR41 pKa = 11.84IADD44 pKa = 3.54YY45 pKa = 10.96LADD48 pKa = 4.84DD49 pKa = 4.12YY50 pKa = 11.66EE51 pKa = 4.96IGYY54 pKa = 9.46VRR56 pKa = 11.84DD57 pKa = 3.71RR58 pKa = 11.84PPKK61 pKa = 7.61HH62 pKa = 6.13RR63 pKa = 11.84RR64 pKa = 11.84YY65 pKa = 10.28AVLRR69 pKa = 11.84TQALFNKK76 pKa = 10.12LIEE79 pKa = 4.47IYY81 pKa = 10.89GKK83 pKa = 10.22LFKK86 pKa = 10.93QNWEE90 pKa = 3.91FLRR93 pKa = 11.84RR94 pKa = 11.84SIDD97 pKa = 3.26GFTTAVGIRR106 pKa = 11.84EE107 pKa = 4.24RR108 pKa = 11.84QWEE111 pKa = 4.18NAARR115 pKa = 11.84AYY117 pKa = 9.43ISCWFFDD124 pKa = 3.95LYY126 pKa = 11.3VSVRR130 pKa = 11.84NAVEE134 pKa = 4.35KK135 pKa = 11.01VSGTAFTQYY144 pKa = 9.92YY145 pKa = 8.29HH146 pKa = 6.59AHH148 pKa = 6.6AAHH151 pKa = 7.12ILHH154 pKa = 7.08EE155 pKa = 4.27YY156 pKa = 10.23DD157 pKa = 4.0ALLVHH162 pKa = 6.82LNAIIRR168 pKa = 11.84PTHH171 pKa = 6.06IKK173 pKa = 10.3LALEE177 pKa = 3.99DD178 pKa = 4.55CIYY181 pKa = 10.29IPRR184 pKa = 11.84IATNMNITNANPFGLAEE201 pKa = 4.26ATWDD205 pKa = 3.51LDD207 pKa = 3.48ADD209 pKa = 4.36FAYY212 pKa = 11.02SIMDD216 pKa = 3.41RR217 pKa = 11.84MKK219 pKa = 10.88ASGSRR224 pKa = 11.84WRR226 pKa = 11.84VEE228 pKa = 3.77PLVTDD233 pKa = 3.69TLGRR237 pKa = 11.84PGWLFDD243 pKa = 3.24WHH245 pKa = 6.22QVGAQQQAYY254 pKa = 9.48AWFPSEE260 pKa = 4.46GNYY263 pKa = 11.0SMEE266 pKa = 4.8DD267 pKa = 3.88LIAPHH272 pKa = 7.14IIGEE276 pKa = 4.25ALTPLMGPCDD286 pKa = 3.9IDD288 pKa = 3.18DD289 pKa = 3.49WQYY292 pKa = 10.44FPNFIRR298 pKa = 11.84PANVPWQNMEE308 pKa = 3.96RR309 pKa = 11.84VSARR313 pKa = 11.84RR314 pKa = 11.84FRR316 pKa = 11.84GSAEE320 pKa = 3.47VRR322 pKa = 11.84TYY324 pKa = 11.4AEE326 pKa = 4.04EE327 pKa = 4.17KK328 pKa = 10.54VNLTRR333 pKa = 11.84FASTGHH339 pKa = 5.23EE340 pKa = 4.11TSKK343 pKa = 10.67KK344 pKa = 8.96RR345 pKa = 11.84SKK347 pKa = 10.47PSSTTTTSSQGGSRR361 pKa = 11.84DD362 pKa = 3.34MEE364 pKa = 4.46TEE366 pKa = 3.94DD367 pKa = 3.67EE368 pKa = 4.2PRR370 pKa = 11.84EE371 pKa = 4.1VVTDD375 pKa = 3.55ITLSQVTSTQQLQTLMTTGEE395 pKa = 3.97ADD397 pKa = 3.54RR398 pKa = 11.84FRR400 pKa = 11.84IIDD403 pKa = 3.52YY404 pKa = 8.86TYY406 pKa = 8.07YY407 pKa = 10.73CCVILRR413 pKa = 11.84NTAQKK418 pKa = 10.21RR419 pKa = 11.84DD420 pKa = 3.13GAIRR424 pKa = 11.84SFCFKK429 pKa = 10.54QQ430 pKa = 3.07
Molecular weight: 49.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
908 |
430 |
478 |
454.0 |
51.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.819 ± 0.848 | 1.432 ± 0.025 |
5.947 ± 0.225 | 6.278 ± 0.472 |
4.295 ± 0.074 | 5.947 ± 1.037 |
2.643 ± 0.215 | 7.489 ± 0.663 |
4.405 ± 0.307 | 7.599 ± 0.896 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.762 ± 0.225 | 3.524 ± 0.024 |
5.176 ± 0.117 | 3.744 ± 0.615 |
7.048 ± 0.425 | 6.167 ± 0.076 |
6.608 ± 1.512 | 5.176 ± 0.356 |
1.762 ± 0.382 | 5.176 ± 0.356 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
