
Torque teno midi virus 9
Taxonomy: Viruses; Anelloviridae; Gammatorquevirus
Average proteome isoelectric point is 7.33
Get precalculated fractions of proteins
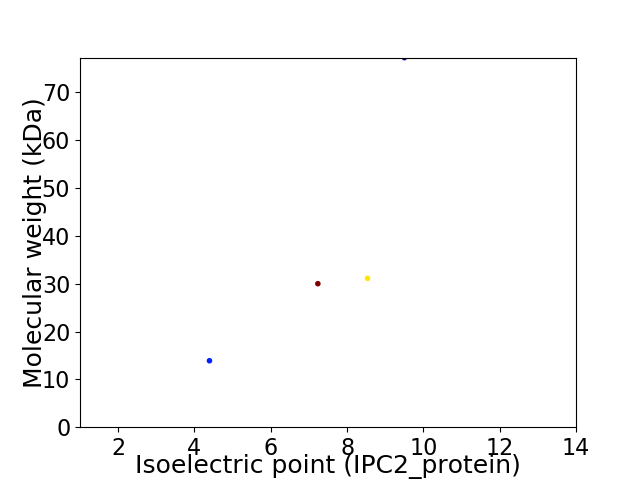
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A7VLY4|A7VLY4_9VIRU Capsid protein OS=Torque teno midi virus 9 OX=2065050 PE=3 SV=1
MM1 pKa = 6.26TAYY4 pKa = 11.0YY5 pKa = 10.7NFARR9 pKa = 11.84KK10 pKa = 7.37TPYY13 pKa = 10.49NNCTHH18 pKa = 5.76EE19 pKa = 4.21QLWISSFNDD28 pKa = 2.9SHH30 pKa = 7.75DD31 pKa = 3.99AFCGCTKK38 pKa = 10.28PVTHH42 pKa = 6.68MLSILFPEE50 pKa = 4.56GHH52 pKa = 7.15RR53 pKa = 11.84DD54 pKa = 3.12LDD56 pKa = 3.59LTIRR60 pKa = 11.84QILQRR65 pKa = 11.84EE66 pKa = 4.19KK67 pKa = 11.01EE68 pKa = 3.98DD69 pKa = 4.02TICLFGGTDD78 pKa = 3.33EE79 pKa = 5.23RR80 pKa = 11.84DD81 pKa = 3.29GGEE84 pKa = 4.02AQEE87 pKa = 5.31DD88 pKa = 4.03PGTKK92 pKa = 10.07EE93 pKa = 4.32KK94 pKa = 10.68DD95 pKa = 3.16IPDD98 pKa = 3.48ATEE101 pKa = 3.82EE102 pKa = 4.35EE103 pKa = 4.46FTKK106 pKa = 10.26IDD108 pKa = 4.37VEE110 pKa = 4.42DD111 pKa = 4.31LLAAAADD118 pKa = 3.87ADD120 pKa = 4.2TPRR123 pKa = 5.39
MM1 pKa = 6.26TAYY4 pKa = 11.0YY5 pKa = 10.7NFARR9 pKa = 11.84KK10 pKa = 7.37TPYY13 pKa = 10.49NNCTHH18 pKa = 5.76EE19 pKa = 4.21QLWISSFNDD28 pKa = 2.9SHH30 pKa = 7.75DD31 pKa = 3.99AFCGCTKK38 pKa = 10.28PVTHH42 pKa = 6.68MLSILFPEE50 pKa = 4.56GHH52 pKa = 7.15RR53 pKa = 11.84DD54 pKa = 3.12LDD56 pKa = 3.59LTIRR60 pKa = 11.84QILQRR65 pKa = 11.84EE66 pKa = 4.19KK67 pKa = 11.01EE68 pKa = 3.98DD69 pKa = 4.02TICLFGGTDD78 pKa = 3.33EE79 pKa = 5.23RR80 pKa = 11.84DD81 pKa = 3.29GGEE84 pKa = 4.02AQEE87 pKa = 5.31DD88 pKa = 4.03PGTKK92 pKa = 10.07EE93 pKa = 4.32KK94 pKa = 10.68DD95 pKa = 3.16IPDD98 pKa = 3.48ATEE101 pKa = 3.82EE102 pKa = 4.35EE103 pKa = 4.46FTKK106 pKa = 10.26IDD108 pKa = 4.37VEE110 pKa = 4.42DD111 pKa = 4.31LLAAAADD118 pKa = 3.87ADD120 pKa = 4.2TPRR123 pKa = 5.39
Molecular weight: 13.9 kDa
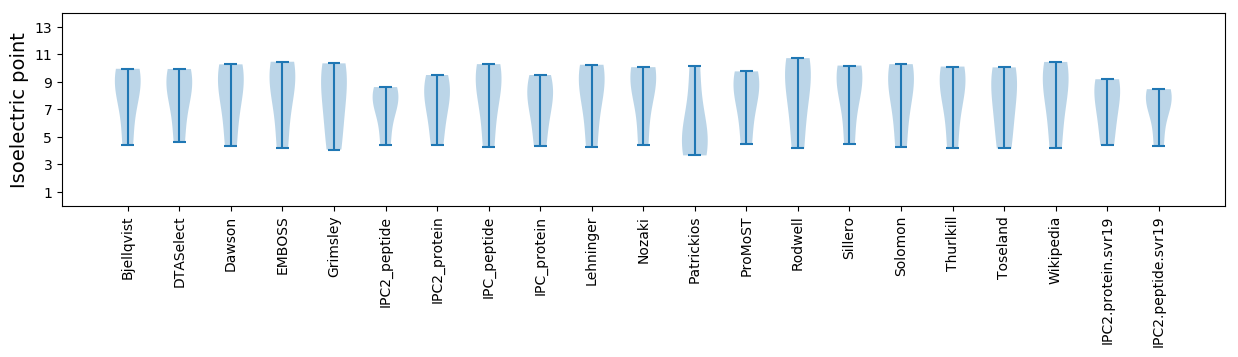
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A7VLY4|A7VLY4_9VIRU Capsid protein OS=Torque teno midi virus 9 OX=2065050 PE=3 SV=1
MM1 pKa = 7.42PFWWNRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 10.02RR10 pKa = 11.84WWRR13 pKa = 11.84GARR16 pKa = 11.84RR17 pKa = 11.84PWYY20 pKa = 9.28KK21 pKa = 9.87RR22 pKa = 11.84KK23 pKa = 10.09RR24 pKa = 11.84YY25 pKa = 5.49TRR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 8.79RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84IYY34 pKa = 10.06KK35 pKa = 9.4NRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84PTRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84HH50 pKa = 4.09TKK52 pKa = 8.95VRR54 pKa = 11.84RR55 pKa = 11.84KK56 pKa = 9.79RR57 pKa = 11.84QTIPVRR63 pKa = 11.84QWQPEE68 pKa = 4.08RR69 pKa = 11.84IVRR72 pKa = 11.84CKK74 pKa = 10.24IKK76 pKa = 10.45GISVIVLGGEE86 pKa = 4.59GKK88 pKa = 9.84QFQCYY93 pKa = 7.34TNEE96 pKa = 3.94ATTWTPLRR104 pKa = 11.84TPGGGGFGVEE114 pKa = 4.91KK115 pKa = 9.97YY116 pKa = 8.82TLSHH120 pKa = 7.24LYY122 pKa = 10.15DD123 pKa = 3.17QYY125 pKa = 12.11KK126 pKa = 9.93FGNNIWTKK134 pKa = 9.71TNILTDD140 pKa = 3.74LVQYY144 pKa = 9.32LGCKK148 pKa = 9.34FVFYY152 pKa = 10.28RR153 pKa = 11.84HH154 pKa = 5.74QYY156 pKa = 8.78VDD158 pKa = 4.79FIVKK162 pKa = 9.78YY163 pKa = 10.62SRR165 pKa = 11.84NLPMTIEE172 pKa = 4.24KK173 pKa = 8.92LTYY176 pKa = 9.93PHH178 pKa = 6.53THH180 pKa = 6.21PQALLLSRR188 pKa = 11.84HH189 pKa = 5.48KK190 pKa = 10.52RR191 pKa = 11.84IIPSRR196 pKa = 11.84VTKK199 pKa = 9.59PHH201 pKa = 5.51GKK203 pKa = 9.07NRR205 pKa = 11.84VIIKK209 pKa = 10.03IKK211 pKa = 7.5PTRR214 pKa = 11.84VVLKK218 pKa = 10.05KK219 pKa = 9.81WFFQEE224 pKa = 4.32TFDD227 pKa = 3.95TTGLCSIYY235 pKa = 10.81AAACDD240 pKa = 4.15LNYY243 pKa = 10.27PHH245 pKa = 7.52IGATSQNQIITLFGLNQSFYY265 pKa = 11.7SKK267 pKa = 10.85GNWGAVVQTGYY278 pKa = 10.61QPYY281 pKa = 9.97PSVTTFTYY289 pKa = 8.76KK290 pKa = 9.83TAATKK295 pKa = 10.09QVEE298 pKa = 4.75TFNKK302 pKa = 6.69PTTYY306 pKa = 9.65EE307 pKa = 3.76ASVSYY312 pKa = 7.61TTGWFNPKK320 pKa = 9.79LMQLTEE326 pKa = 4.29YY327 pKa = 8.73VTPQQLAKK335 pKa = 9.93PVLAFRR341 pKa = 11.84YY342 pKa = 8.66NAQLDD347 pKa = 3.78NGKK350 pKa = 10.67GNSVWFKK357 pKa = 11.3SILTEE362 pKa = 4.48DD363 pKa = 3.82FSKK366 pKa = 11.05PKK368 pKa = 10.16VDD370 pKa = 3.42SDD372 pKa = 5.43LIISDD377 pKa = 3.65MPIWQALYY385 pKa = 10.84GFTDD389 pKa = 4.95FIKK392 pKa = 10.6AVKK395 pKa = 9.58PDD397 pKa = 3.95PNFLASYY404 pKa = 9.94ILLIEE409 pKa = 4.26TRR411 pKa = 11.84FVEE414 pKa = 4.66PFSSHH419 pKa = 4.97ATGNYY424 pKa = 7.61WIPIDD429 pKa = 3.85SSFVTGKK436 pKa = 10.71GPYY439 pKa = 7.45NTPIGDD445 pKa = 3.61YY446 pKa = 10.38TKK448 pKa = 10.26TKK450 pKa = 9.64WFPTVKK456 pKa = 10.25NQQEE460 pKa = 4.5SINAFVKK467 pKa = 10.38AGPYY471 pKa = 9.48IPRR474 pKa = 11.84LQDD477 pKa = 3.22EE478 pKa = 4.94KK479 pKa = 11.18KK480 pKa = 9.56STWEE484 pKa = 3.37LHH486 pKa = 6.38AFYY489 pKa = 10.53QFYY492 pKa = 10.58LKK494 pKa = 10.27WGGTYY499 pKa = 9.08TEE501 pKa = 4.38EE502 pKa = 4.16TQVADD507 pKa = 3.83PSKK510 pKa = 10.02QGTYY514 pKa = 10.93VMPSFIQEE522 pKa = 3.89KK523 pKa = 10.03LQIINPAKK531 pKa = 10.05QKK533 pKa = 10.3AAAMLHH539 pKa = 5.63CWDD542 pKa = 3.89YY543 pKa = 11.43RR544 pKa = 11.84RR545 pKa = 11.84GLITQSAIKK554 pKa = 9.72RR555 pKa = 11.84ICEE558 pKa = 4.0DD559 pKa = 3.42QPTDD563 pKa = 3.11TDD565 pKa = 3.93FQTGSEE571 pKa = 4.19SSQPLKK577 pKa = 10.64KK578 pKa = 10.04KK579 pKa = 10.63KK580 pKa = 9.1KK581 pKa = 4.62TTKK584 pKa = 9.79EE585 pKa = 3.47LPYY588 pKa = 9.98PPKK591 pKa = 10.53EE592 pKa = 3.97KK593 pKa = 11.0EE594 pKa = 4.04EE595 pKa = 3.98LQKK598 pKa = 11.11CLLSLYY604 pKa = 9.95EE605 pKa = 3.95EE606 pKa = 5.22SISQEE611 pKa = 3.53IPQTQNLQEE620 pKa = 5.42LIQQQQQQQQQLKK633 pKa = 9.22YY634 pKa = 9.43NILKK638 pKa = 10.2IISEE642 pKa = 4.64LKK644 pKa = 9.95QQQQLLGLQTGLVLL658 pKa = 4.97
MM1 pKa = 7.42PFWWNRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 10.02RR10 pKa = 11.84WWRR13 pKa = 11.84GARR16 pKa = 11.84RR17 pKa = 11.84PWYY20 pKa = 9.28KK21 pKa = 9.87RR22 pKa = 11.84KK23 pKa = 10.09RR24 pKa = 11.84YY25 pKa = 5.49TRR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 8.79RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84IYY34 pKa = 10.06KK35 pKa = 9.4NRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84PTRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84HH50 pKa = 4.09TKK52 pKa = 8.95VRR54 pKa = 11.84RR55 pKa = 11.84KK56 pKa = 9.79RR57 pKa = 11.84QTIPVRR63 pKa = 11.84QWQPEE68 pKa = 4.08RR69 pKa = 11.84IVRR72 pKa = 11.84CKK74 pKa = 10.24IKK76 pKa = 10.45GISVIVLGGEE86 pKa = 4.59GKK88 pKa = 9.84QFQCYY93 pKa = 7.34TNEE96 pKa = 3.94ATTWTPLRR104 pKa = 11.84TPGGGGFGVEE114 pKa = 4.91KK115 pKa = 9.97YY116 pKa = 8.82TLSHH120 pKa = 7.24LYY122 pKa = 10.15DD123 pKa = 3.17QYY125 pKa = 12.11KK126 pKa = 9.93FGNNIWTKK134 pKa = 9.71TNILTDD140 pKa = 3.74LVQYY144 pKa = 9.32LGCKK148 pKa = 9.34FVFYY152 pKa = 10.28RR153 pKa = 11.84HH154 pKa = 5.74QYY156 pKa = 8.78VDD158 pKa = 4.79FIVKK162 pKa = 9.78YY163 pKa = 10.62SRR165 pKa = 11.84NLPMTIEE172 pKa = 4.24KK173 pKa = 8.92LTYY176 pKa = 9.93PHH178 pKa = 6.53THH180 pKa = 6.21PQALLLSRR188 pKa = 11.84HH189 pKa = 5.48KK190 pKa = 10.52RR191 pKa = 11.84IIPSRR196 pKa = 11.84VTKK199 pKa = 9.59PHH201 pKa = 5.51GKK203 pKa = 9.07NRR205 pKa = 11.84VIIKK209 pKa = 10.03IKK211 pKa = 7.5PTRR214 pKa = 11.84VVLKK218 pKa = 10.05KK219 pKa = 9.81WFFQEE224 pKa = 4.32TFDD227 pKa = 3.95TTGLCSIYY235 pKa = 10.81AAACDD240 pKa = 4.15LNYY243 pKa = 10.27PHH245 pKa = 7.52IGATSQNQIITLFGLNQSFYY265 pKa = 11.7SKK267 pKa = 10.85GNWGAVVQTGYY278 pKa = 10.61QPYY281 pKa = 9.97PSVTTFTYY289 pKa = 8.76KK290 pKa = 9.83TAATKK295 pKa = 10.09QVEE298 pKa = 4.75TFNKK302 pKa = 6.69PTTYY306 pKa = 9.65EE307 pKa = 3.76ASVSYY312 pKa = 7.61TTGWFNPKK320 pKa = 9.79LMQLTEE326 pKa = 4.29YY327 pKa = 8.73VTPQQLAKK335 pKa = 9.93PVLAFRR341 pKa = 11.84YY342 pKa = 8.66NAQLDD347 pKa = 3.78NGKK350 pKa = 10.67GNSVWFKK357 pKa = 11.3SILTEE362 pKa = 4.48DD363 pKa = 3.82FSKK366 pKa = 11.05PKK368 pKa = 10.16VDD370 pKa = 3.42SDD372 pKa = 5.43LIISDD377 pKa = 3.65MPIWQALYY385 pKa = 10.84GFTDD389 pKa = 4.95FIKK392 pKa = 10.6AVKK395 pKa = 9.58PDD397 pKa = 3.95PNFLASYY404 pKa = 9.94ILLIEE409 pKa = 4.26TRR411 pKa = 11.84FVEE414 pKa = 4.66PFSSHH419 pKa = 4.97ATGNYY424 pKa = 7.61WIPIDD429 pKa = 3.85SSFVTGKK436 pKa = 10.71GPYY439 pKa = 7.45NTPIGDD445 pKa = 3.61YY446 pKa = 10.38TKK448 pKa = 10.26TKK450 pKa = 9.64WFPTVKK456 pKa = 10.25NQQEE460 pKa = 4.5SINAFVKK467 pKa = 10.38AGPYY471 pKa = 9.48IPRR474 pKa = 11.84LQDD477 pKa = 3.22EE478 pKa = 4.94KK479 pKa = 11.18KK480 pKa = 9.56STWEE484 pKa = 3.37LHH486 pKa = 6.38AFYY489 pKa = 10.53QFYY492 pKa = 10.58LKK494 pKa = 10.27WGGTYY499 pKa = 9.08TEE501 pKa = 4.38EE502 pKa = 4.16TQVADD507 pKa = 3.83PSKK510 pKa = 10.02QGTYY514 pKa = 10.93VMPSFIQEE522 pKa = 3.89KK523 pKa = 10.03LQIINPAKK531 pKa = 10.05QKK533 pKa = 10.3AAAMLHH539 pKa = 5.63CWDD542 pKa = 3.89YY543 pKa = 11.43RR544 pKa = 11.84RR545 pKa = 11.84GLITQSAIKK554 pKa = 9.72RR555 pKa = 11.84ICEE558 pKa = 4.0DD559 pKa = 3.42QPTDD563 pKa = 3.11TDD565 pKa = 3.93FQTGSEE571 pKa = 4.19SSQPLKK577 pKa = 10.64KK578 pKa = 10.04KK579 pKa = 10.63KK580 pKa = 9.1KK581 pKa = 4.62TTKK584 pKa = 9.79EE585 pKa = 3.47LPYY588 pKa = 9.98PPKK591 pKa = 10.53EE592 pKa = 3.97KK593 pKa = 11.0EE594 pKa = 4.04EE595 pKa = 3.98LQKK598 pKa = 11.11CLLSLYY604 pKa = 9.95EE605 pKa = 3.95EE606 pKa = 5.22SISQEE611 pKa = 3.53IPQTQNLQEE620 pKa = 5.42LIQQQQQQQQQLKK633 pKa = 9.22YY634 pKa = 9.43NILKK638 pKa = 10.2IISEE642 pKa = 4.64LKK644 pKa = 9.95QQQQLLGLQTGLVLL658 pKa = 4.97
Molecular weight: 77.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1322 |
123 |
658 |
330.5 |
38.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.749 ± 1.154 | 1.891 ± 0.422 |
4.917 ± 1.194 | 5.825 ± 0.921 |
4.236 ± 0.405 | 4.539 ± 0.575 |
2.572 ± 0.67 | 5.295 ± 0.826 |
8.623 ± 0.825 | 7.337 ± 0.193 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.059 ± 0.096 | 4.085 ± 0.249 |
5.976 ± 0.869 | 6.581 ± 0.989 |
6.203 ± 0.773 | 6.732 ± 2.464 |
9.228 ± 0.594 | 3.48 ± 0.749 |
1.664 ± 0.688 | 4.009 ± 1.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
