
Miniopterus schreibersii polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
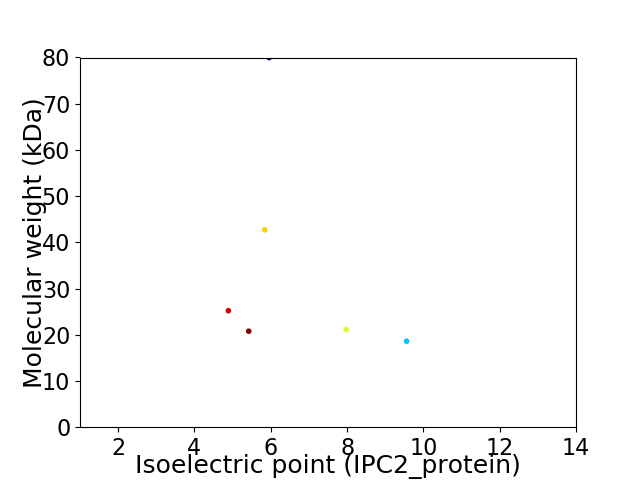
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1S7J007|A0A1S7J007_9POLY Small T antigen OS=Miniopterus schreibersii polyomavirus 1 OX=1904408 PE=4 SV=1
MM1 pKa = 7.45GGVLSTVVEE10 pKa = 4.84MITMAVEE17 pKa = 3.9LSAATGIAMDD27 pKa = 4.35ALLTGEE33 pKa = 4.62ALAALEE39 pKa = 4.48AEE41 pKa = 4.87VTSLMTIQGLSAFEE55 pKa = 4.56ALAQLGWTAEE65 pKa = 4.19QFSNMAFISTTFSQAIGYY83 pKa = 9.53GVMFQTVTGISGLVQAGIRR102 pKa = 11.84LGLNVASSSRR112 pKa = 11.84GTQVAQLEE120 pKa = 4.28RR121 pKa = 11.84DD122 pKa = 3.25FGKK125 pKa = 10.2IVEE128 pKa = 4.42TLHH131 pKa = 6.82VNLSHH136 pKa = 6.94QMNPLNWCASLHH148 pKa = 5.86EE149 pKa = 4.8EE150 pKa = 4.42FPPSIQHH157 pKa = 6.19LPISLRR163 pKa = 11.84HH164 pKa = 5.2QFADD168 pKa = 3.64LLKK171 pKa = 10.49LGRR174 pKa = 11.84WVNQHH179 pKa = 6.37HH180 pKa = 6.4FTTNPAFEE188 pKa = 4.89SGDD191 pKa = 3.39VIRR194 pKa = 11.84RR195 pKa = 11.84FEE197 pKa = 4.22APGGAFQDD205 pKa = 4.04VAPDD209 pKa = 3.37WLVNLILRR217 pKa = 11.84LHH219 pKa = 7.04DD220 pKa = 3.93GAEE223 pKa = 4.07EE224 pKa = 3.91ATPFCSSLSSS234 pKa = 3.35
MM1 pKa = 7.45GGVLSTVVEE10 pKa = 4.84MITMAVEE17 pKa = 3.9LSAATGIAMDD27 pKa = 4.35ALLTGEE33 pKa = 4.62ALAALEE39 pKa = 4.48AEE41 pKa = 4.87VTSLMTIQGLSAFEE55 pKa = 4.56ALAQLGWTAEE65 pKa = 4.19QFSNMAFISTTFSQAIGYY83 pKa = 9.53GVMFQTVTGISGLVQAGIRR102 pKa = 11.84LGLNVASSSRR112 pKa = 11.84GTQVAQLEE120 pKa = 4.28RR121 pKa = 11.84DD122 pKa = 3.25FGKK125 pKa = 10.2IVEE128 pKa = 4.42TLHH131 pKa = 6.82VNLSHH136 pKa = 6.94QMNPLNWCASLHH148 pKa = 5.86EE149 pKa = 4.8EE150 pKa = 4.42FPPSIQHH157 pKa = 6.19LPISLRR163 pKa = 11.84HH164 pKa = 5.2QFADD168 pKa = 3.64LLKK171 pKa = 10.49LGRR174 pKa = 11.84WVNQHH179 pKa = 6.37HH180 pKa = 6.4FTTNPAFEE188 pKa = 4.89SGDD191 pKa = 3.39VIRR194 pKa = 11.84RR195 pKa = 11.84FEE197 pKa = 4.22APGGAFQDD205 pKa = 4.04VAPDD209 pKa = 3.37WLVNLILRR217 pKa = 11.84LHH219 pKa = 7.04DD220 pKa = 3.93GAEE223 pKa = 4.07EE224 pKa = 3.91ATPFCSSLSSS234 pKa = 3.35
Molecular weight: 25.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S7J009|A0A1S7J009_9POLY Putative alternative large T antigen OS=Miniopterus schreibersii polyomavirus 1 OX=1904408 PE=4 SV=1
MM1 pKa = 8.07DD2 pKa = 5.46PPASRR7 pKa = 11.84KK8 pKa = 8.17STGHH12 pKa = 5.73GRR14 pKa = 11.84QEE16 pKa = 4.0YY17 pKa = 10.93LPMKK21 pKa = 10.03DD22 pKa = 4.26LNPNQIYY29 pKa = 8.28TVMNPVTPTVEE40 pKa = 4.44SPPSHH45 pKa = 7.08PLNPQVITLLPPAEE59 pKa = 4.95DD60 pKa = 3.63SQPPPLPPKK69 pKa = 8.76RR70 pKa = 11.84TPPRR74 pKa = 11.84RR75 pKa = 11.84SVSLPSRR82 pKa = 11.84KK83 pKa = 8.92PLPMHH88 pKa = 6.69PLQMEE93 pKa = 4.41VEE95 pKa = 4.25TLEE98 pKa = 4.09RR99 pKa = 11.84NLRR102 pKa = 11.84QKK104 pKa = 9.44EE105 pKa = 3.82QDD107 pKa = 3.58MVRR110 pKa = 11.84MWMDD114 pKa = 3.47LVQARR119 pKa = 11.84KK120 pKa = 7.98QASRR124 pKa = 11.84ARR126 pKa = 11.84LQRR129 pKa = 11.84KK130 pKa = 5.9KK131 pKa = 10.61QKK133 pKa = 9.2WVTVLLICLLVFLSILAMLYY153 pKa = 10.83LLIKK157 pKa = 10.73LLTTSS162 pKa = 3.42
MM1 pKa = 8.07DD2 pKa = 5.46PPASRR7 pKa = 11.84KK8 pKa = 8.17STGHH12 pKa = 5.73GRR14 pKa = 11.84QEE16 pKa = 4.0YY17 pKa = 10.93LPMKK21 pKa = 10.03DD22 pKa = 4.26LNPNQIYY29 pKa = 8.28TVMNPVTPTVEE40 pKa = 4.44SPPSHH45 pKa = 7.08PLNPQVITLLPPAEE59 pKa = 4.95DD60 pKa = 3.63SQPPPLPPKK69 pKa = 8.76RR70 pKa = 11.84TPPRR74 pKa = 11.84RR75 pKa = 11.84SVSLPSRR82 pKa = 11.84KK83 pKa = 8.92PLPMHH88 pKa = 6.69PLQMEE93 pKa = 4.41VEE95 pKa = 4.25TLEE98 pKa = 4.09RR99 pKa = 11.84NLRR102 pKa = 11.84QKK104 pKa = 9.44EE105 pKa = 3.82QDD107 pKa = 3.58MVRR110 pKa = 11.84MWMDD114 pKa = 3.47LVQARR119 pKa = 11.84KK120 pKa = 7.98QASRR124 pKa = 11.84ARR126 pKa = 11.84LQRR129 pKa = 11.84KK130 pKa = 5.9KK131 pKa = 10.61QKK133 pKa = 9.2WVTVLLICLLVFLSILAMLYY153 pKa = 10.83LLIKK157 pKa = 10.73LLTTSS162 pKa = 3.42
Molecular weight: 18.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1870 |
162 |
710 |
311.7 |
34.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.059 ± 0.794 | 2.62 ± 0.774 |
4.118 ± 0.396 | 6.31 ± 0.588 |
4.652 ± 0.749 | 5.989 ± 1.042 |
2.513 ± 0.349 | 4.064 ± 0.557 |
6.043 ± 1.38 | 11.818 ± 0.636 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.674 ± 0.402 | 3.904 ± 0.384 |
6.684 ± 1.44 | 5.027 ± 0.573 |
3.957 ± 0.535 | 7.326 ± 0.62 |
5.775 ± 0.25 | 5.829 ± 0.437 |
1.444 ± 0.379 | 2.193 ± 0.505 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
