
Beihai rhabdo-like virus 5
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Nyamiviridae; Berhavirus; Echinoderm berhavirus
Average proteome isoelectric point is 7.57
Get precalculated fractions of proteins
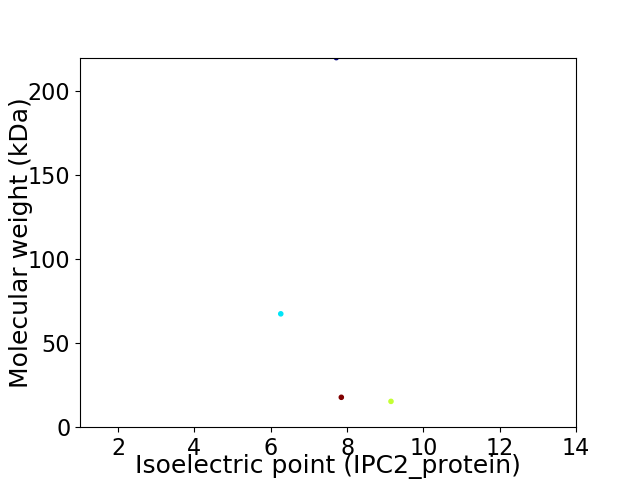
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KMR4|A0A1L3KMR4_9MONO Uncharacterized protein OS=Beihai rhabdo-like virus 5 OX=1922655 PE=4 SV=1
MM1 pKa = 7.18GHH3 pKa = 7.12DD4 pKa = 4.48LSHH7 pKa = 7.79ISIFSFQKK15 pKa = 10.37ADD17 pKa = 3.44LVQDD21 pKa = 3.45YY22 pKa = 11.28GNVVKK27 pKa = 10.73KK28 pKa = 9.82ATGRR32 pKa = 11.84ARR34 pKa = 11.84TSNMTGNRR42 pKa = 11.84KK43 pKa = 8.02TINTVPYY50 pKa = 9.3IQMPSPLMEE59 pKa = 3.87QAFVCHH65 pKa = 6.77KK66 pKa = 6.96MTLRR70 pKa = 11.84LGEE73 pKa = 5.03EE74 pKa = 3.7EE75 pKa = 4.59AEE77 pKa = 4.26WYY79 pKa = 9.98VPRR82 pKa = 11.84PAGCSDD88 pKa = 3.47AEE90 pKa = 4.48YY91 pKa = 10.91GSSAHH96 pKa = 6.24LKK98 pKa = 10.61LVISWSALEE107 pKa = 4.88PILEE111 pKa = 4.18EE112 pKa = 4.06LRR114 pKa = 11.84GSAEE118 pKa = 3.9QQKK121 pKa = 9.94QLAGAAGCTKK131 pKa = 10.25EE132 pKa = 3.87VLEE135 pKa = 4.04ILRR138 pKa = 11.84GFFLISEE145 pKa = 4.62AEE147 pKa = 4.31VATLWAHH154 pKa = 6.65HH155 pKa = 5.66EE156 pKa = 4.63AIHH159 pKa = 5.81GAAMEE164 pKa = 4.11QGEE167 pKa = 4.5PEE169 pKa = 4.07ATLGGSPGKK178 pKa = 10.11KK179 pKa = 9.15RR180 pKa = 11.84PRR182 pKa = 11.84KK183 pKa = 8.89EE184 pKa = 3.75KK185 pKa = 9.84PQASPEE191 pKa = 3.91QSEE194 pKa = 4.49EE195 pKa = 4.17EE196 pKa = 4.32EE197 pKa = 4.31ATSGPRR203 pKa = 11.84RR204 pKa = 11.84KK205 pKa = 9.82RR206 pKa = 11.84FRR208 pKa = 11.84RR209 pKa = 11.84KK210 pKa = 8.55GTTKK214 pKa = 10.46FGRR217 pKa = 11.84FAAFKK222 pKa = 10.7AKK224 pKa = 10.85YY225 pKa = 9.2EE226 pKa = 4.24DD227 pKa = 4.48ANHH230 pKa = 6.77PMYY233 pKa = 10.29QKK235 pKa = 10.91KK236 pKa = 9.84ILLEE240 pKa = 4.21GADD243 pKa = 3.77RR244 pKa = 11.84EE245 pKa = 4.49IYY247 pKa = 9.72QATIYY252 pKa = 9.93SHH254 pKa = 7.11PNVAFAEE261 pKa = 4.25YY262 pKa = 9.48CYY264 pKa = 11.26AKK266 pKa = 9.43ATQSRR271 pKa = 11.84GVEE274 pKa = 4.17GQKK277 pKa = 10.35KK278 pKa = 5.32EE279 pKa = 3.95TRR281 pKa = 11.84FAMGNFILTCLAAFLPGVLFSNAKK305 pKa = 9.95FDD307 pKa = 3.66DD308 pKa = 4.02RR309 pKa = 11.84QYY311 pKa = 11.85GSYY314 pKa = 10.53QLGPVLKK321 pKa = 9.88EE322 pKa = 3.83GSTTEE327 pKa = 3.79QEE329 pKa = 4.05QYY331 pKa = 10.34EE332 pKa = 4.51WPEE335 pKa = 3.62LTAAASRR342 pKa = 11.84ARR344 pKa = 11.84YY345 pKa = 9.84LEE347 pKa = 3.69ILQIGFYY354 pKa = 10.57AALACVKK361 pKa = 10.52SVTQEE366 pKa = 3.68NLSFLEE372 pKa = 4.09KK373 pKa = 9.94RR374 pKa = 11.84WNAAISQSDD383 pKa = 3.73LTSPSKK389 pKa = 10.16TNDD392 pKa = 2.98FSFDD396 pKa = 3.65PANLISFSTNAGDD409 pKa = 3.9YY410 pKa = 10.59LSKK413 pKa = 10.89SDD415 pKa = 5.43LYY417 pKa = 11.29FLTSSLLAEE426 pKa = 4.16TDD428 pKa = 3.35EE429 pKa = 5.54AEE431 pKa = 4.38TSGSIASCLLKK442 pKa = 10.11QVEE445 pKa = 4.85MICEE449 pKa = 3.95YY450 pKa = 11.24SGMPRR455 pKa = 11.84IKK457 pKa = 10.95LMMNWATRR465 pKa = 11.84FDD467 pKa = 4.16SSAHH471 pKa = 5.14YY472 pKa = 9.53CHH474 pKa = 6.75AVVTEE479 pKa = 4.72IISFLTILQEE489 pKa = 3.91IQAKK493 pKa = 10.14VSTAEE498 pKa = 4.05FPFLRR503 pKa = 11.84LTRR506 pKa = 11.84PALLEE511 pKa = 4.06RR512 pKa = 11.84LSARR516 pKa = 11.84SFPHH520 pKa = 6.54LCFCATMDD528 pKa = 4.78AKK530 pKa = 8.76MTKK533 pKa = 9.91NLAVGYY539 pKa = 9.63AGKK542 pKa = 10.0DD543 pKa = 3.49FPMTMSAKK551 pKa = 10.32RR552 pKa = 11.84LQTLLRR558 pKa = 11.84QKK560 pKa = 10.47IPDD563 pKa = 3.46VGALSEE569 pKa = 4.33VQVAALASLGIQVEE583 pKa = 4.07GDD585 pKa = 3.13IDD587 pKa = 3.9EE588 pKa = 4.99VIQVARR594 pKa = 11.84KK595 pKa = 9.42RR596 pKa = 11.84KK597 pKa = 7.96RR598 pKa = 11.84QLVEE602 pKa = 3.86SDD604 pKa = 3.03SDD606 pKa = 3.49
MM1 pKa = 7.18GHH3 pKa = 7.12DD4 pKa = 4.48LSHH7 pKa = 7.79ISIFSFQKK15 pKa = 10.37ADD17 pKa = 3.44LVQDD21 pKa = 3.45YY22 pKa = 11.28GNVVKK27 pKa = 10.73KK28 pKa = 9.82ATGRR32 pKa = 11.84ARR34 pKa = 11.84TSNMTGNRR42 pKa = 11.84KK43 pKa = 8.02TINTVPYY50 pKa = 9.3IQMPSPLMEE59 pKa = 3.87QAFVCHH65 pKa = 6.77KK66 pKa = 6.96MTLRR70 pKa = 11.84LGEE73 pKa = 5.03EE74 pKa = 3.7EE75 pKa = 4.59AEE77 pKa = 4.26WYY79 pKa = 9.98VPRR82 pKa = 11.84PAGCSDD88 pKa = 3.47AEE90 pKa = 4.48YY91 pKa = 10.91GSSAHH96 pKa = 6.24LKK98 pKa = 10.61LVISWSALEE107 pKa = 4.88PILEE111 pKa = 4.18EE112 pKa = 4.06LRR114 pKa = 11.84GSAEE118 pKa = 3.9QQKK121 pKa = 9.94QLAGAAGCTKK131 pKa = 10.25EE132 pKa = 3.87VLEE135 pKa = 4.04ILRR138 pKa = 11.84GFFLISEE145 pKa = 4.62AEE147 pKa = 4.31VATLWAHH154 pKa = 6.65HH155 pKa = 5.66EE156 pKa = 4.63AIHH159 pKa = 5.81GAAMEE164 pKa = 4.11QGEE167 pKa = 4.5PEE169 pKa = 4.07ATLGGSPGKK178 pKa = 10.11KK179 pKa = 9.15RR180 pKa = 11.84PRR182 pKa = 11.84KK183 pKa = 8.89EE184 pKa = 3.75KK185 pKa = 9.84PQASPEE191 pKa = 3.91QSEE194 pKa = 4.49EE195 pKa = 4.17EE196 pKa = 4.32EE197 pKa = 4.31ATSGPRR203 pKa = 11.84RR204 pKa = 11.84KK205 pKa = 9.82RR206 pKa = 11.84FRR208 pKa = 11.84RR209 pKa = 11.84KK210 pKa = 8.55GTTKK214 pKa = 10.46FGRR217 pKa = 11.84FAAFKK222 pKa = 10.7AKK224 pKa = 10.85YY225 pKa = 9.2EE226 pKa = 4.24DD227 pKa = 4.48ANHH230 pKa = 6.77PMYY233 pKa = 10.29QKK235 pKa = 10.91KK236 pKa = 9.84ILLEE240 pKa = 4.21GADD243 pKa = 3.77RR244 pKa = 11.84EE245 pKa = 4.49IYY247 pKa = 9.72QATIYY252 pKa = 9.93SHH254 pKa = 7.11PNVAFAEE261 pKa = 4.25YY262 pKa = 9.48CYY264 pKa = 11.26AKK266 pKa = 9.43ATQSRR271 pKa = 11.84GVEE274 pKa = 4.17GQKK277 pKa = 10.35KK278 pKa = 5.32EE279 pKa = 3.95TRR281 pKa = 11.84FAMGNFILTCLAAFLPGVLFSNAKK305 pKa = 9.95FDD307 pKa = 3.66DD308 pKa = 4.02RR309 pKa = 11.84QYY311 pKa = 11.85GSYY314 pKa = 10.53QLGPVLKK321 pKa = 9.88EE322 pKa = 3.83GSTTEE327 pKa = 3.79QEE329 pKa = 4.05QYY331 pKa = 10.34EE332 pKa = 4.51WPEE335 pKa = 3.62LTAAASRR342 pKa = 11.84ARR344 pKa = 11.84YY345 pKa = 9.84LEE347 pKa = 3.69ILQIGFYY354 pKa = 10.57AALACVKK361 pKa = 10.52SVTQEE366 pKa = 3.68NLSFLEE372 pKa = 4.09KK373 pKa = 9.94RR374 pKa = 11.84WNAAISQSDD383 pKa = 3.73LTSPSKK389 pKa = 10.16TNDD392 pKa = 2.98FSFDD396 pKa = 3.65PANLISFSTNAGDD409 pKa = 3.9YY410 pKa = 10.59LSKK413 pKa = 10.89SDD415 pKa = 5.43LYY417 pKa = 11.29FLTSSLLAEE426 pKa = 4.16TDD428 pKa = 3.35EE429 pKa = 5.54AEE431 pKa = 4.38TSGSIASCLLKK442 pKa = 10.11QVEE445 pKa = 4.85MICEE449 pKa = 3.95YY450 pKa = 11.24SGMPRR455 pKa = 11.84IKK457 pKa = 10.95LMMNWATRR465 pKa = 11.84FDD467 pKa = 4.16SSAHH471 pKa = 5.14YY472 pKa = 9.53CHH474 pKa = 6.75AVVTEE479 pKa = 4.72IISFLTILQEE489 pKa = 3.91IQAKK493 pKa = 10.14VSTAEE498 pKa = 4.05FPFLRR503 pKa = 11.84LTRR506 pKa = 11.84PALLEE511 pKa = 4.06RR512 pKa = 11.84LSARR516 pKa = 11.84SFPHH520 pKa = 6.54LCFCATMDD528 pKa = 4.78AKK530 pKa = 8.76MTKK533 pKa = 9.91NLAVGYY539 pKa = 9.63AGKK542 pKa = 10.0DD543 pKa = 3.49FPMTMSAKK551 pKa = 10.32RR552 pKa = 11.84LQTLLRR558 pKa = 11.84QKK560 pKa = 10.47IPDD563 pKa = 3.46VGALSEE569 pKa = 4.33VQVAALASLGIQVEE583 pKa = 4.07GDD585 pKa = 3.13IDD587 pKa = 3.9EE588 pKa = 4.99VIQVARR594 pKa = 11.84KK595 pKa = 9.42RR596 pKa = 11.84KK597 pKa = 7.96RR598 pKa = 11.84QLVEE602 pKa = 3.86SDD604 pKa = 3.03SDD606 pKa = 3.49
Molecular weight: 67.41 kDa
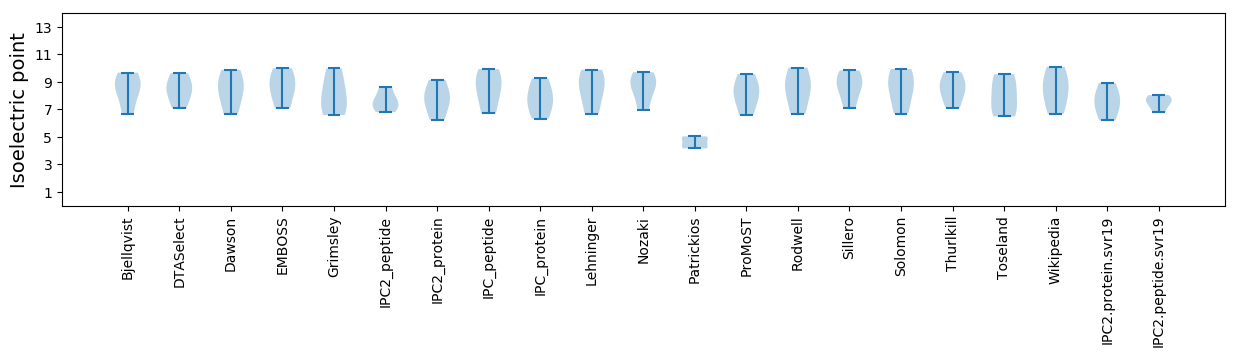
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KMP2|A0A1L3KMP2_9MONO Putative nucleoprotein OS=Beihai rhabdo-like virus 5 OX=1922655 PE=4 SV=1
MM1 pKa = 7.51EE2 pKa = 4.52VFLLFILLGNCLGRR16 pKa = 11.84KK17 pKa = 8.98RR18 pKa = 11.84RR19 pKa = 11.84FEE21 pKa = 3.99FMVSIGHH28 pKa = 5.7TEE30 pKa = 3.74QSYY33 pKa = 8.15MCHH36 pKa = 7.48DD37 pKa = 3.18IGKK40 pKa = 9.83LYY42 pKa = 10.3RR43 pKa = 11.84HH44 pKa = 6.59RR45 pKa = 11.84FNRR48 pKa = 11.84MNANPKK54 pKa = 8.84VSFYY58 pKa = 10.66IPEE61 pKa = 4.3SVLGEE66 pKa = 4.05QAAHH70 pKa = 6.79FDD72 pKa = 3.96LDD74 pKa = 3.73SDD76 pKa = 4.16CEE78 pKa = 4.16YY79 pKa = 11.12FPVEE83 pKa = 3.9QSGIPAYY90 pKa = 9.31WFHH93 pKa = 6.67SVQRR97 pKa = 11.84PKK99 pKa = 10.12IQYY102 pKa = 9.08RR103 pKa = 11.84GYY105 pKa = 9.32MLKK108 pKa = 10.22KK109 pKa = 8.72PHH111 pKa = 5.29GTRR114 pKa = 11.84GRR116 pKa = 11.84FKK118 pKa = 10.83HH119 pKa = 5.17QFEE122 pKa = 4.31LFARR126 pKa = 11.84RR127 pKa = 11.84RR128 pKa = 3.61
MM1 pKa = 7.51EE2 pKa = 4.52VFLLFILLGNCLGRR16 pKa = 11.84KK17 pKa = 8.98RR18 pKa = 11.84RR19 pKa = 11.84FEE21 pKa = 3.99FMVSIGHH28 pKa = 5.7TEE30 pKa = 3.74QSYY33 pKa = 8.15MCHH36 pKa = 7.48DD37 pKa = 3.18IGKK40 pKa = 9.83LYY42 pKa = 10.3RR43 pKa = 11.84HH44 pKa = 6.59RR45 pKa = 11.84FNRR48 pKa = 11.84MNANPKK54 pKa = 8.84VSFYY58 pKa = 10.66IPEE61 pKa = 4.3SVLGEE66 pKa = 4.05QAAHH70 pKa = 6.79FDD72 pKa = 3.96LDD74 pKa = 3.73SDD76 pKa = 4.16CEE78 pKa = 4.16YY79 pKa = 11.12FPVEE83 pKa = 3.9QSGIPAYY90 pKa = 9.31WFHH93 pKa = 6.67SVQRR97 pKa = 11.84PKK99 pKa = 10.12IQYY102 pKa = 9.08RR103 pKa = 11.84GYY105 pKa = 9.32MLKK108 pKa = 10.22KK109 pKa = 8.72PHH111 pKa = 5.29GTRR114 pKa = 11.84GRR116 pKa = 11.84FKK118 pKa = 10.83HH119 pKa = 5.17QFEE122 pKa = 4.31LFARR126 pKa = 11.84RR127 pKa = 11.84RR128 pKa = 3.61
Molecular weight: 15.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2812 |
128 |
1920 |
703.0 |
80.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.504 ± 1.329 | 2.027 ± 0.283 |
4.765 ± 0.532 | 6.508 ± 0.824 |
4.659 ± 0.559 | 4.943 ± 0.586 |
3.058 ± 0.538 | 5.192 ± 0.233 |
5.832 ± 0.512 | 10.242 ± 0.529 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.24 ± 0.302 | 3.734 ± 0.589 |
4.232 ± 0.315 | 4.125 ± 0.376 |
6.472 ± 0.606 | 7.361 ± 0.944 |
6.294 ± 0.599 | 5.583 ± 0.536 |
0.96 ± 0.194 | 4.267 ± 0.515 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
