
Polaribacter sp. IC066
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Polaribacter; unclassified Polaribacter
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
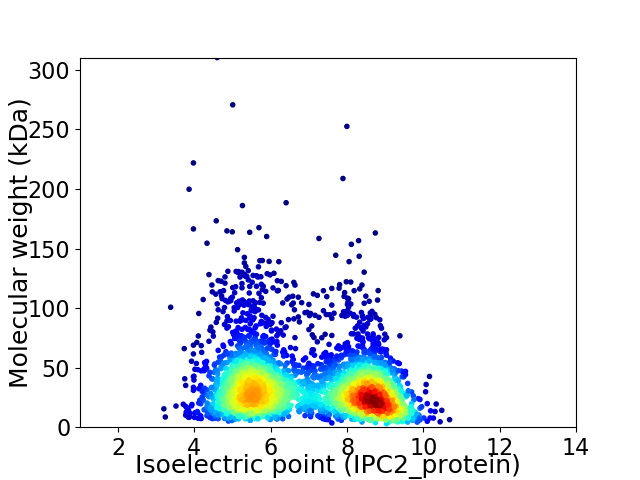
Virtual 2D-PAGE plot for 3473 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C6YBI1|A0A5C6YBI1_9FLAO Molecular chaperone DjiA OS=Polaribacter sp. IC066 OX=57032 GN=ES044_05975 PE=4 SV=1
MM1 pKa = 7.51TKK3 pKa = 10.66LKK5 pKa = 10.38FLFVALFCAALVLQTTSCSTNDD27 pKa = 3.89DD28 pKa = 3.93GPGGEE33 pKa = 4.16EE34 pKa = 3.92VGNDD38 pKa = 3.39VQITGISINPTSLSLEE54 pKa = 4.03EE55 pKa = 4.92GVTSNLVVSLEE66 pKa = 4.08PSNATGTVTWSSSNPAVATVDD87 pKa = 3.1SDD89 pKa = 4.05GLVSSISEE97 pKa = 4.13GTASIAAAIGTFSSVCDD114 pKa = 3.38VTVTKK119 pKa = 10.39EE120 pKa = 3.74AVIVDD125 pKa = 5.01DD126 pKa = 4.03ITLKK130 pKa = 10.99GSDD133 pKa = 3.97YY134 pKa = 11.26YY135 pKa = 10.34IIQLDD140 pKa = 3.94EE141 pKa = 4.78DD142 pKa = 4.27SYY144 pKa = 11.96SAIEE148 pKa = 4.2SKK150 pKa = 10.68VIQDD154 pKa = 3.92FRR156 pKa = 11.84PDD158 pKa = 3.39DD159 pKa = 3.72TTKK162 pKa = 10.66FLYY165 pKa = 10.37VWDD168 pKa = 3.99GTFEE172 pKa = 4.42GGNSVGNNFYY182 pKa = 11.16GLDD185 pKa = 3.71QDD187 pKa = 4.36WISLKK192 pKa = 10.46VGNVGWSGAGFSVGSAYY209 pKa = 10.56GDD211 pKa = 2.95IDD213 pKa = 4.0MTPMFDD219 pKa = 4.01NPEE222 pKa = 4.04DD223 pKa = 4.14YY224 pKa = 10.73YY225 pKa = 11.77LHH227 pKa = 7.24IGFKK231 pKa = 9.43TGQSASSYY239 pKa = 11.16LFILTDD245 pKa = 3.85GQAEE249 pKa = 4.07AKK251 pKa = 10.0IAIGNDD257 pKa = 3.17FNDD260 pKa = 4.73DD261 pKa = 3.31GTTYY265 pKa = 10.69TSYY268 pKa = 11.22KK269 pKa = 10.08QLTRR273 pKa = 11.84DD274 pKa = 3.73NTWNSIDD281 pKa = 4.29IPVTHH286 pKa = 7.27LNQLGLFYY294 pKa = 10.89NQTFQDD300 pKa = 3.76VNIFTMLAGGNQGTTLDD317 pKa = 3.44MDD319 pKa = 4.05AVFFYY324 pKa = 10.93KK325 pKa = 10.47KK326 pKa = 10.62
MM1 pKa = 7.51TKK3 pKa = 10.66LKK5 pKa = 10.38FLFVALFCAALVLQTTSCSTNDD27 pKa = 3.89DD28 pKa = 3.93GPGGEE33 pKa = 4.16EE34 pKa = 3.92VGNDD38 pKa = 3.39VQITGISINPTSLSLEE54 pKa = 4.03EE55 pKa = 4.92GVTSNLVVSLEE66 pKa = 4.08PSNATGTVTWSSSNPAVATVDD87 pKa = 3.1SDD89 pKa = 4.05GLVSSISEE97 pKa = 4.13GTASIAAAIGTFSSVCDD114 pKa = 3.38VTVTKK119 pKa = 10.39EE120 pKa = 3.74AVIVDD125 pKa = 5.01DD126 pKa = 4.03ITLKK130 pKa = 10.99GSDD133 pKa = 3.97YY134 pKa = 11.26YY135 pKa = 10.34IIQLDD140 pKa = 3.94EE141 pKa = 4.78DD142 pKa = 4.27SYY144 pKa = 11.96SAIEE148 pKa = 4.2SKK150 pKa = 10.68VIQDD154 pKa = 3.92FRR156 pKa = 11.84PDD158 pKa = 3.39DD159 pKa = 3.72TTKK162 pKa = 10.66FLYY165 pKa = 10.37VWDD168 pKa = 3.99GTFEE172 pKa = 4.42GGNSVGNNFYY182 pKa = 11.16GLDD185 pKa = 3.71QDD187 pKa = 4.36WISLKK192 pKa = 10.46VGNVGWSGAGFSVGSAYY209 pKa = 10.56GDD211 pKa = 2.95IDD213 pKa = 4.0MTPMFDD219 pKa = 4.01NPEE222 pKa = 4.04DD223 pKa = 4.14YY224 pKa = 10.73YY225 pKa = 11.77LHH227 pKa = 7.24IGFKK231 pKa = 9.43TGQSASSYY239 pKa = 11.16LFILTDD245 pKa = 3.85GQAEE249 pKa = 4.07AKK251 pKa = 10.0IAIGNDD257 pKa = 3.17FNDD260 pKa = 4.73DD261 pKa = 3.31GTTYY265 pKa = 10.69TSYY268 pKa = 11.22KK269 pKa = 10.08QLTRR273 pKa = 11.84DD274 pKa = 3.73NTWNSIDD281 pKa = 4.29IPVTHH286 pKa = 7.27LNQLGLFYY294 pKa = 10.89NQTFQDD300 pKa = 3.76VNIFTMLAGGNQGTTLDD317 pKa = 3.44MDD319 pKa = 4.05AVFFYY324 pKa = 10.93KK325 pKa = 10.47KK326 pKa = 10.62
Molecular weight: 35.08 kDa
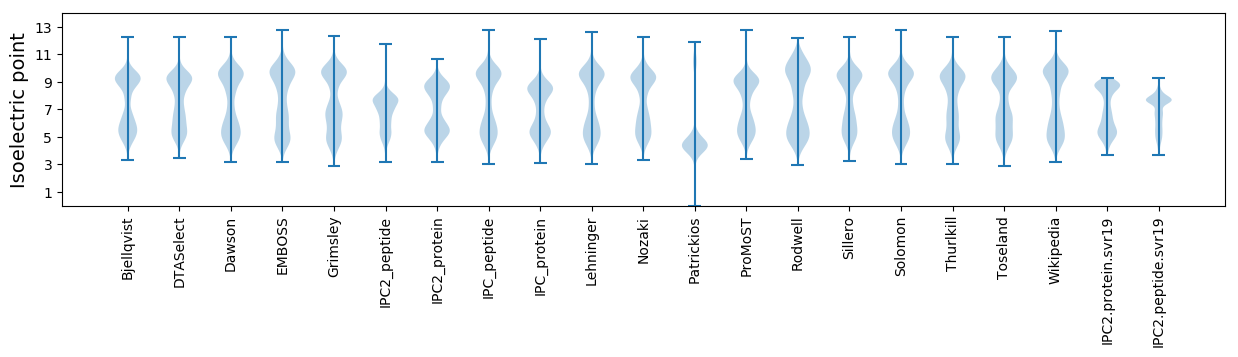
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C6YES7|A0A5C6YES7_9FLAO TonB-dependent receptor OS=Polaribacter sp. IC066 OX=57032 GN=ES044_02925 PE=3 SV=1
MM1 pKa = 7.56KK2 pKa = 10.32KK3 pKa = 10.58GIFILLGMLLMVSTIEE19 pKa = 4.22AKK21 pKa = 10.46KK22 pKa = 10.34SQNTANNFSFTYY34 pKa = 10.09AYY36 pKa = 10.8NNAVNFIEE44 pKa = 4.34NGVEE48 pKa = 4.71FFIFTNGDD56 pKa = 2.91FDD58 pKa = 5.92FDD60 pKa = 3.36TRR62 pKa = 11.84TNDD65 pKa = 2.72RR66 pKa = 11.84RR67 pKa = 11.84VRR69 pKa = 11.84IHH71 pKa = 7.09RR72 pKa = 11.84DD73 pKa = 2.98FNGRR77 pKa = 11.84IRR79 pKa = 11.84SIGNVHH85 pKa = 6.71LRR87 pKa = 11.84YY88 pKa = 9.93DD89 pKa = 3.28LRR91 pKa = 11.84GNVTRR96 pKa = 11.84IGAISIRR103 pKa = 11.84YY104 pKa = 8.14FRR106 pKa = 11.84GRR108 pKa = 11.84LTNVGDD114 pKa = 3.38LRR116 pKa = 11.84VRR118 pKa = 11.84YY119 pKa = 9.24NRR121 pKa = 11.84WEE123 pKa = 3.97YY124 pKa = 10.09PVFYY128 pKa = 10.92GNVRR132 pKa = 11.84NFYY135 pKa = 9.39YY136 pKa = 11.02NNGVRR141 pKa = 11.84FNVSFGDD148 pKa = 3.17ICNYY152 pKa = 9.57NDD154 pKa = 3.22TYY156 pKa = 11.07FFRR159 pKa = 11.84NDD161 pKa = 2.9FSRR164 pKa = 11.84NYY166 pKa = 8.03TQIRR170 pKa = 11.84EE171 pKa = 4.12DD172 pKa = 3.59RR173 pKa = 11.84NFYY176 pKa = 10.58YY177 pKa = 10.87YY178 pKa = 10.07QANPNARR185 pKa = 11.84IGKK188 pKa = 9.67RR189 pKa = 11.84STILKK194 pKa = 8.71RR195 pKa = 11.84RR196 pKa = 11.84KK197 pKa = 9.31PISVNNNNNRR207 pKa = 11.84RR208 pKa = 11.84KK209 pKa = 10.35NSATSSNNSYY219 pKa = 10.61RR220 pKa = 11.84KK221 pKa = 8.77PQRR224 pKa = 11.84TTNTRR229 pKa = 11.84NTAIKK234 pKa = 10.33RR235 pKa = 11.84STRR238 pKa = 11.84NSNTTSKK245 pKa = 9.8TDD247 pKa = 3.51ADD249 pKa = 4.01RR250 pKa = 11.84TPAIKK255 pKa = 9.53TEE257 pKa = 3.89RR258 pKa = 11.84EE259 pKa = 3.89TLKK262 pKa = 10.41EE263 pKa = 3.54RR264 pKa = 11.84RR265 pKa = 11.84SFKK268 pKa = 10.62KK269 pKa = 9.13STRR272 pKa = 11.84NEE274 pKa = 3.86AKK276 pKa = 9.78TLEE279 pKa = 4.28STRR282 pKa = 11.84NKK284 pKa = 10.72YY285 pKa = 7.47EE286 pKa = 3.84VKK288 pKa = 9.76TKK290 pKa = 9.8SRR292 pKa = 11.84RR293 pKa = 11.84NTRR296 pKa = 11.84NKK298 pKa = 10.21SDD300 pKa = 3.27RR301 pKa = 11.84SS302 pKa = 3.6
MM1 pKa = 7.56KK2 pKa = 10.32KK3 pKa = 10.58GIFILLGMLLMVSTIEE19 pKa = 4.22AKK21 pKa = 10.46KK22 pKa = 10.34SQNTANNFSFTYY34 pKa = 10.09AYY36 pKa = 10.8NNAVNFIEE44 pKa = 4.34NGVEE48 pKa = 4.71FFIFTNGDD56 pKa = 2.91FDD58 pKa = 5.92FDD60 pKa = 3.36TRR62 pKa = 11.84TNDD65 pKa = 2.72RR66 pKa = 11.84RR67 pKa = 11.84VRR69 pKa = 11.84IHH71 pKa = 7.09RR72 pKa = 11.84DD73 pKa = 2.98FNGRR77 pKa = 11.84IRR79 pKa = 11.84SIGNVHH85 pKa = 6.71LRR87 pKa = 11.84YY88 pKa = 9.93DD89 pKa = 3.28LRR91 pKa = 11.84GNVTRR96 pKa = 11.84IGAISIRR103 pKa = 11.84YY104 pKa = 8.14FRR106 pKa = 11.84GRR108 pKa = 11.84LTNVGDD114 pKa = 3.38LRR116 pKa = 11.84VRR118 pKa = 11.84YY119 pKa = 9.24NRR121 pKa = 11.84WEE123 pKa = 3.97YY124 pKa = 10.09PVFYY128 pKa = 10.92GNVRR132 pKa = 11.84NFYY135 pKa = 9.39YY136 pKa = 11.02NNGVRR141 pKa = 11.84FNVSFGDD148 pKa = 3.17ICNYY152 pKa = 9.57NDD154 pKa = 3.22TYY156 pKa = 11.07FFRR159 pKa = 11.84NDD161 pKa = 2.9FSRR164 pKa = 11.84NYY166 pKa = 8.03TQIRR170 pKa = 11.84EE171 pKa = 4.12DD172 pKa = 3.59RR173 pKa = 11.84NFYY176 pKa = 10.58YY177 pKa = 10.87YY178 pKa = 10.07QANPNARR185 pKa = 11.84IGKK188 pKa = 9.67RR189 pKa = 11.84STILKK194 pKa = 8.71RR195 pKa = 11.84RR196 pKa = 11.84KK197 pKa = 9.31PISVNNNNNRR207 pKa = 11.84RR208 pKa = 11.84KK209 pKa = 10.35NSATSSNNSYY219 pKa = 10.61RR220 pKa = 11.84KK221 pKa = 8.77PQRR224 pKa = 11.84TTNTRR229 pKa = 11.84NTAIKK234 pKa = 10.33RR235 pKa = 11.84STRR238 pKa = 11.84NSNTTSKK245 pKa = 9.8TDD247 pKa = 3.51ADD249 pKa = 4.01RR250 pKa = 11.84TPAIKK255 pKa = 9.53TEE257 pKa = 3.89RR258 pKa = 11.84EE259 pKa = 3.89TLKK262 pKa = 10.41EE263 pKa = 3.54RR264 pKa = 11.84RR265 pKa = 11.84SFKK268 pKa = 10.62KK269 pKa = 9.13STRR272 pKa = 11.84NEE274 pKa = 3.86AKK276 pKa = 9.78TLEE279 pKa = 4.28STRR282 pKa = 11.84NKK284 pKa = 10.72YY285 pKa = 7.47EE286 pKa = 3.84VKK288 pKa = 9.76TKK290 pKa = 9.8SRR292 pKa = 11.84RR293 pKa = 11.84NTRR296 pKa = 11.84NKK298 pKa = 10.21SDD300 pKa = 3.27RR301 pKa = 11.84SS302 pKa = 3.6
Molecular weight: 35.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1147573 |
24 |
2805 |
330.4 |
37.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.268 ± 0.039 | 0.689 ± 0.011 |
5.326 ± 0.028 | 6.594 ± 0.045 |
5.548 ± 0.029 | 6.057 ± 0.043 |
1.693 ± 0.018 | 8.472 ± 0.042 |
8.635 ± 0.05 | 9.252 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.069 ± 0.02 | 6.508 ± 0.049 |
3.027 ± 0.02 | 3.299 ± 0.022 |
3.213 ± 0.024 | 6.553 ± 0.031 |
5.889 ± 0.032 | 6.031 ± 0.032 |
0.995 ± 0.015 | 3.881 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
