
Ursus maritimus papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Omegapapillomavirus; Omegapapillomavirus 1
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
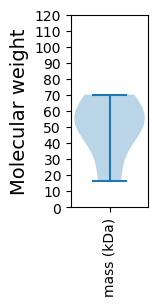
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|B2KKW9|B2KKW9_9PAPI Regulatory protein E2 OS=Ursus maritimus papillomavirus 1 OX=461322 GN=E2 PE=3 SV=1
MM1 pKa = 7.85AEE3 pKa = 4.56RR4 pKa = 11.84PGSTPGGEE12 pKa = 4.34YY13 pKa = 9.85ILRR16 pKa = 11.84EE17 pKa = 4.33AEE19 pKa = 4.43CSDD22 pKa = 3.47SSTDD26 pKa = 3.87GEE28 pKa = 4.43GEE30 pKa = 4.01EE31 pKa = 4.57GNEE34 pKa = 4.47DD35 pKa = 3.5EE36 pKa = 6.49GIDD39 pKa = 4.26FIDD42 pKa = 5.58DD43 pKa = 3.73ASQCIGHH50 pKa = 5.85EE51 pKa = 4.0HH52 pKa = 6.7LALFACQTRR61 pKa = 11.84QADD64 pKa = 4.04SEE66 pKa = 4.39HH67 pKa = 6.11LQQLKK72 pKa = 10.14RR73 pKa = 11.84KK74 pKa = 8.24YY75 pKa = 10.14ASPSPSPKK83 pKa = 9.9QKK85 pKa = 10.61VNSDD89 pKa = 3.75CALSPRR95 pKa = 11.84LAAISLSQEE104 pKa = 3.57RR105 pKa = 11.84KK106 pKa = 8.43AKK108 pKa = 10.06RR109 pKa = 11.84RR110 pKa = 11.84LFAALPDD117 pKa = 3.66SGYY120 pKa = 11.35GNTLEE125 pKa = 5.18AEE127 pKa = 4.09AEE129 pKa = 4.13NRR131 pKa = 11.84VEE133 pKa = 4.03EE134 pKa = 4.15QTQVEE139 pKa = 4.52SSGEE143 pKa = 3.7EE144 pKa = 3.78MARR147 pKa = 11.84NGSSSNLNLRR157 pKa = 11.84NAAIPIQLVRR167 pKa = 11.84SKK169 pKa = 11.57DD170 pKa = 3.53SMLTKK175 pKa = 10.26LAKK178 pKa = 10.16FKK180 pKa = 10.47EE181 pKa = 4.56VYY183 pKa = 10.19DD184 pKa = 3.99ISFKK188 pKa = 11.27DD189 pKa = 3.12LTRR192 pKa = 11.84PFKK195 pKa = 10.79SSKK198 pKa = 8.47TCNSDD203 pKa = 2.44WVVAVFGVHH212 pKa = 6.22EE213 pKa = 4.75SVCEE217 pKa = 3.98GSIEE221 pKa = 4.13LLKK224 pKa = 10.69KK225 pKa = 9.42HH226 pKa = 5.75CTYY229 pKa = 11.14VHH231 pKa = 6.02LQKK234 pKa = 10.69EE235 pKa = 4.33LSSWGNVLLLLLCFTTAKK253 pKa = 10.49NRR255 pKa = 11.84TTVEE259 pKa = 3.98KK260 pKa = 10.56LLCSVLACNQQLLMADD276 pKa = 3.71PPKK279 pKa = 10.71VRR281 pKa = 11.84SAAAAVYY288 pKa = 8.84WYY290 pKa = 10.53KK291 pKa = 11.19AGFTNHH297 pKa = 4.8ATQWGEE303 pKa = 3.71KK304 pKa = 8.92PKK306 pKa = 10.27WIMQHH311 pKa = 6.23VGLQHH316 pKa = 6.68IAADD320 pKa = 3.9QIPFDD325 pKa = 4.65LSVMIQWAYY334 pKa = 11.01DD335 pKa = 3.12NGYY338 pKa = 10.1QDD340 pKa = 3.9EE341 pKa = 4.9CEE343 pKa = 4.17VAYY346 pKa = 10.09EE347 pKa = 3.99YY348 pKa = 11.56ARR350 pKa = 11.84LADD353 pKa = 3.59TDD355 pKa = 4.14TNASAFLKK363 pKa = 10.68SNCQAKK369 pKa = 9.48YY370 pKa = 10.94VKK372 pKa = 10.51DD373 pKa = 3.74CVTMVRR379 pKa = 11.84LYY381 pKa = 10.73KK382 pKa = 9.97RR383 pKa = 11.84AEE385 pKa = 3.79MSRR388 pKa = 11.84LNMRR392 pKa = 11.84QWIEE396 pKa = 3.72KK397 pKa = 9.91RR398 pKa = 11.84RR399 pKa = 11.84EE400 pKa = 3.92EE401 pKa = 4.51VDD403 pKa = 3.81GDD405 pKa = 4.07GDD407 pKa = 3.1WRR409 pKa = 11.84TIVTFLKK416 pKa = 8.71YY417 pKa = 10.51QNVEE421 pKa = 4.34FVPFMQKK428 pKa = 10.12FKK430 pKa = 10.39MLLKK434 pKa = 10.5GVPKK438 pKa = 9.55KK439 pKa = 10.75NCMVFVGPPNTGKK452 pKa = 10.34SAFCMGLLKK461 pKa = 10.57FMQGRR466 pKa = 11.84VISFMNSRR474 pKa = 11.84SHH476 pKa = 7.18FWIQPLQDD484 pKa = 3.75AKK486 pKa = 11.23LGLLDD491 pKa = 5.49DD492 pKa = 4.65ATDD495 pKa = 3.82ACWDD499 pKa = 3.61FFDD502 pKa = 4.72VYY504 pKa = 10.33MRR506 pKa = 11.84NALDD510 pKa = 4.12GNPISIDD517 pKa = 3.58RR518 pKa = 11.84KK519 pKa = 9.57HH520 pKa = 6.41KK521 pKa = 10.96APIQMKK527 pKa = 9.97CPPLLVTTNVDD538 pKa = 3.1VTNNDD543 pKa = 2.39RR544 pKa = 11.84WKK546 pKa = 9.02YY547 pKa = 8.38LHH549 pKa = 6.66SRR551 pKa = 11.84LVTVRR556 pKa = 11.84FPNEE560 pKa = 3.55FPITDD565 pKa = 3.64EE566 pKa = 4.59GEE568 pKa = 4.18VVYY571 pKa = 10.86KK572 pKa = 9.32FTEE575 pKa = 4.23QNWKK579 pKa = 10.65SFFGRR584 pKa = 11.84LWARR588 pKa = 11.84LDD590 pKa = 3.66LSDD593 pKa = 5.26GEE595 pKa = 4.58EE596 pKa = 4.25EE597 pKa = 4.81GDD599 pKa = 4.38DD600 pKa = 5.32AEE602 pKa = 4.33PQQPFRR608 pKa = 11.84CTARR612 pKa = 11.84SDD614 pKa = 3.77DD615 pKa = 4.07GYY617 pKa = 11.77LL618 pKa = 3.41
MM1 pKa = 7.85AEE3 pKa = 4.56RR4 pKa = 11.84PGSTPGGEE12 pKa = 4.34YY13 pKa = 9.85ILRR16 pKa = 11.84EE17 pKa = 4.33AEE19 pKa = 4.43CSDD22 pKa = 3.47SSTDD26 pKa = 3.87GEE28 pKa = 4.43GEE30 pKa = 4.01EE31 pKa = 4.57GNEE34 pKa = 4.47DD35 pKa = 3.5EE36 pKa = 6.49GIDD39 pKa = 4.26FIDD42 pKa = 5.58DD43 pKa = 3.73ASQCIGHH50 pKa = 5.85EE51 pKa = 4.0HH52 pKa = 6.7LALFACQTRR61 pKa = 11.84QADD64 pKa = 4.04SEE66 pKa = 4.39HH67 pKa = 6.11LQQLKK72 pKa = 10.14RR73 pKa = 11.84KK74 pKa = 8.24YY75 pKa = 10.14ASPSPSPKK83 pKa = 9.9QKK85 pKa = 10.61VNSDD89 pKa = 3.75CALSPRR95 pKa = 11.84LAAISLSQEE104 pKa = 3.57RR105 pKa = 11.84KK106 pKa = 8.43AKK108 pKa = 10.06RR109 pKa = 11.84RR110 pKa = 11.84LFAALPDD117 pKa = 3.66SGYY120 pKa = 11.35GNTLEE125 pKa = 5.18AEE127 pKa = 4.09AEE129 pKa = 4.13NRR131 pKa = 11.84VEE133 pKa = 4.03EE134 pKa = 4.15QTQVEE139 pKa = 4.52SSGEE143 pKa = 3.7EE144 pKa = 3.78MARR147 pKa = 11.84NGSSSNLNLRR157 pKa = 11.84NAAIPIQLVRR167 pKa = 11.84SKK169 pKa = 11.57DD170 pKa = 3.53SMLTKK175 pKa = 10.26LAKK178 pKa = 10.16FKK180 pKa = 10.47EE181 pKa = 4.56VYY183 pKa = 10.19DD184 pKa = 3.99ISFKK188 pKa = 11.27DD189 pKa = 3.12LTRR192 pKa = 11.84PFKK195 pKa = 10.79SSKK198 pKa = 8.47TCNSDD203 pKa = 2.44WVVAVFGVHH212 pKa = 6.22EE213 pKa = 4.75SVCEE217 pKa = 3.98GSIEE221 pKa = 4.13LLKK224 pKa = 10.69KK225 pKa = 9.42HH226 pKa = 5.75CTYY229 pKa = 11.14VHH231 pKa = 6.02LQKK234 pKa = 10.69EE235 pKa = 4.33LSSWGNVLLLLLCFTTAKK253 pKa = 10.49NRR255 pKa = 11.84TTVEE259 pKa = 3.98KK260 pKa = 10.56LLCSVLACNQQLLMADD276 pKa = 3.71PPKK279 pKa = 10.71VRR281 pKa = 11.84SAAAAVYY288 pKa = 8.84WYY290 pKa = 10.53KK291 pKa = 11.19AGFTNHH297 pKa = 4.8ATQWGEE303 pKa = 3.71KK304 pKa = 8.92PKK306 pKa = 10.27WIMQHH311 pKa = 6.23VGLQHH316 pKa = 6.68IAADD320 pKa = 3.9QIPFDD325 pKa = 4.65LSVMIQWAYY334 pKa = 11.01DD335 pKa = 3.12NGYY338 pKa = 10.1QDD340 pKa = 3.9EE341 pKa = 4.9CEE343 pKa = 4.17VAYY346 pKa = 10.09EE347 pKa = 3.99YY348 pKa = 11.56ARR350 pKa = 11.84LADD353 pKa = 3.59TDD355 pKa = 4.14TNASAFLKK363 pKa = 10.68SNCQAKK369 pKa = 9.48YY370 pKa = 10.94VKK372 pKa = 10.51DD373 pKa = 3.74CVTMVRR379 pKa = 11.84LYY381 pKa = 10.73KK382 pKa = 9.97RR383 pKa = 11.84AEE385 pKa = 3.79MSRR388 pKa = 11.84LNMRR392 pKa = 11.84QWIEE396 pKa = 3.72KK397 pKa = 9.91RR398 pKa = 11.84RR399 pKa = 11.84EE400 pKa = 3.92EE401 pKa = 4.51VDD403 pKa = 3.81GDD405 pKa = 4.07GDD407 pKa = 3.1WRR409 pKa = 11.84TIVTFLKK416 pKa = 8.71YY417 pKa = 10.51QNVEE421 pKa = 4.34FVPFMQKK428 pKa = 10.12FKK430 pKa = 10.39MLLKK434 pKa = 10.5GVPKK438 pKa = 9.55KK439 pKa = 10.75NCMVFVGPPNTGKK452 pKa = 10.34SAFCMGLLKK461 pKa = 10.57FMQGRR466 pKa = 11.84VISFMNSRR474 pKa = 11.84SHH476 pKa = 7.18FWIQPLQDD484 pKa = 3.75AKK486 pKa = 11.23LGLLDD491 pKa = 5.49DD492 pKa = 4.65ATDD495 pKa = 3.82ACWDD499 pKa = 3.61FFDD502 pKa = 4.72VYY504 pKa = 10.33MRR506 pKa = 11.84NALDD510 pKa = 4.12GNPISIDD517 pKa = 3.58RR518 pKa = 11.84KK519 pKa = 9.57HH520 pKa = 6.41KK521 pKa = 10.96APIQMKK527 pKa = 9.97CPPLLVTTNVDD538 pKa = 3.1VTNNDD543 pKa = 2.39RR544 pKa = 11.84WKK546 pKa = 9.02YY547 pKa = 8.38LHH549 pKa = 6.66SRR551 pKa = 11.84LVTVRR556 pKa = 11.84FPNEE560 pKa = 3.55FPITDD565 pKa = 3.64EE566 pKa = 4.59GEE568 pKa = 4.18VVYY571 pKa = 10.86KK572 pKa = 9.32FTEE575 pKa = 4.23QNWKK579 pKa = 10.65SFFGRR584 pKa = 11.84LWARR588 pKa = 11.84LDD590 pKa = 3.66LSDD593 pKa = 5.26GEE595 pKa = 4.58EE596 pKa = 4.25EE597 pKa = 4.81GDD599 pKa = 4.38DD600 pKa = 5.32AEE602 pKa = 4.33PQQPFRR608 pKa = 11.84CTARR612 pKa = 11.84SDD614 pKa = 3.77DD615 pKa = 4.07GYY617 pKa = 11.77LL618 pKa = 3.41
Molecular weight: 70.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B2KKX0|B2KKX0_9PAPI Minor capsid protein L2 OS=Ursus maritimus papillomavirus 1 OX=461322 GN=L2 PE=3 SV=1
MM1 pKa = 7.62EE2 pKa = 5.19KK3 pKa = 10.12RR4 pKa = 11.84RR5 pKa = 11.84EE6 pKa = 4.21TTQSLSSRR14 pKa = 11.84LGALQDD20 pKa = 3.42QMMDD24 pKa = 2.84IYY26 pKa = 10.93EE27 pKa = 4.19KK28 pKa = 10.59GGRR31 pKa = 11.84GLEE34 pKa = 4.04VPLMYY39 pKa = 9.62WALQRR44 pKa = 11.84KK45 pKa = 7.32EE46 pKa = 3.98HH47 pKa = 6.35ALLYY51 pKa = 10.23LARR54 pKa = 11.84QNGYY58 pKa = 10.49DD59 pKa = 3.61KK60 pKa = 11.31LGLQPVPALAVSKK73 pKa = 11.03AKK75 pKa = 9.18ATQAISMHH83 pKa = 6.61LLLCSLQDD91 pKa = 3.3SEE93 pKa = 5.27YY94 pKa = 10.94GQEE97 pKa = 3.89PWSLSDD103 pKa = 3.37TSVEE107 pKa = 4.16LYY109 pKa = 10.03EE110 pKa = 5.0SKK112 pKa = 10.17PQKK115 pKa = 9.0TFKK118 pKa = 10.63KK119 pKa = 8.9HH120 pKa = 4.29GATVRR125 pKa = 11.84VHH127 pKa = 6.53YY128 pKa = 10.74DD129 pKa = 3.11NSAEE133 pKa = 3.91NAMEE137 pKa = 4.02YY138 pKa = 10.21VCWGKK143 pKa = 10.66VYY145 pKa = 10.49SWDD148 pKa = 3.56FCSSKK153 pKa = 8.29WTCCTCGVDD162 pKa = 3.7EE163 pKa = 5.58RR164 pKa = 11.84GVYY167 pKa = 9.64VNKK170 pKa = 9.91KK171 pKa = 9.9DD172 pKa = 3.32KK173 pKa = 10.33KK174 pKa = 9.26QYY176 pKa = 10.35YY177 pKa = 10.15EE178 pKa = 4.13NFEE181 pKa = 4.38KK182 pKa = 10.99DD183 pKa = 3.0SVTYY187 pKa = 10.44GKK189 pKa = 8.68TGTWTVQFKK198 pKa = 11.28NKK200 pKa = 8.26TFCCSTSSSVGEE212 pKa = 4.03PVPTASHH219 pKa = 7.08DD220 pKa = 4.03APDD223 pKa = 4.33GGAPCDD229 pKa = 3.65PNSPQKK235 pKa = 10.99AKK237 pKa = 11.01APFSGKK243 pKa = 9.6RR244 pKa = 11.84KK245 pKa = 9.07HH246 pKa = 6.36NGPSVHH252 pKa = 6.4GSEE255 pKa = 5.13ASGPLLGGQGEE266 pKa = 4.62RR267 pKa = 11.84GPSPKK272 pKa = 9.39RR273 pKa = 11.84HH274 pKa = 5.97KK275 pKa = 9.8PASDD279 pKa = 3.59GEE281 pKa = 4.38RR282 pKa = 11.84TLRR285 pKa = 11.84QPTHH289 pKa = 6.42AAHH292 pKa = 6.47PTGTPVSLHH301 pKa = 5.51VQQQQQQHH309 pKa = 4.81QQQQLQQQQLQQLQQQQLQQQQQRR333 pKa = 11.84RR334 pKa = 11.84PQEE337 pKa = 3.98AEE339 pKa = 4.14DD340 pKa = 3.51RR341 pKa = 11.84HH342 pKa = 5.47QPRR345 pKa = 11.84ADD347 pKa = 3.39LPRR350 pKa = 11.84RR351 pKa = 11.84DD352 pKa = 4.31PLHH355 pKa = 6.42PASTPVPVIVLSGSPNQLKK374 pKa = 10.37CYY376 pKa = 10.18RR377 pKa = 11.84YY378 pKa = 10.07RR379 pKa = 11.84LNKK382 pKa = 9.26KK383 pKa = 9.04HH384 pKa = 6.21KK385 pKa = 9.63CLIKK389 pKa = 10.39FVSTTWYY396 pKa = 5.63WTNEE400 pKa = 3.47GGQGEE405 pKa = 4.5KK406 pKa = 11.23AMITLMFYY414 pKa = 10.85SEE416 pKa = 4.16ACKK419 pKa = 9.66NTFLNSVVLPKK430 pKa = 10.81GVTSTGGFLSLL441 pKa = 4.64
MM1 pKa = 7.62EE2 pKa = 5.19KK3 pKa = 10.12RR4 pKa = 11.84RR5 pKa = 11.84EE6 pKa = 4.21TTQSLSSRR14 pKa = 11.84LGALQDD20 pKa = 3.42QMMDD24 pKa = 2.84IYY26 pKa = 10.93EE27 pKa = 4.19KK28 pKa = 10.59GGRR31 pKa = 11.84GLEE34 pKa = 4.04VPLMYY39 pKa = 9.62WALQRR44 pKa = 11.84KK45 pKa = 7.32EE46 pKa = 3.98HH47 pKa = 6.35ALLYY51 pKa = 10.23LARR54 pKa = 11.84QNGYY58 pKa = 10.49DD59 pKa = 3.61KK60 pKa = 11.31LGLQPVPALAVSKK73 pKa = 11.03AKK75 pKa = 9.18ATQAISMHH83 pKa = 6.61LLLCSLQDD91 pKa = 3.3SEE93 pKa = 5.27YY94 pKa = 10.94GQEE97 pKa = 3.89PWSLSDD103 pKa = 3.37TSVEE107 pKa = 4.16LYY109 pKa = 10.03EE110 pKa = 5.0SKK112 pKa = 10.17PQKK115 pKa = 9.0TFKK118 pKa = 10.63KK119 pKa = 8.9HH120 pKa = 4.29GATVRR125 pKa = 11.84VHH127 pKa = 6.53YY128 pKa = 10.74DD129 pKa = 3.11NSAEE133 pKa = 3.91NAMEE137 pKa = 4.02YY138 pKa = 10.21VCWGKK143 pKa = 10.66VYY145 pKa = 10.49SWDD148 pKa = 3.56FCSSKK153 pKa = 8.29WTCCTCGVDD162 pKa = 3.7EE163 pKa = 5.58RR164 pKa = 11.84GVYY167 pKa = 9.64VNKK170 pKa = 9.91KK171 pKa = 9.9DD172 pKa = 3.32KK173 pKa = 10.33KK174 pKa = 9.26QYY176 pKa = 10.35YY177 pKa = 10.15EE178 pKa = 4.13NFEE181 pKa = 4.38KK182 pKa = 10.99DD183 pKa = 3.0SVTYY187 pKa = 10.44GKK189 pKa = 8.68TGTWTVQFKK198 pKa = 11.28NKK200 pKa = 8.26TFCCSTSSSVGEE212 pKa = 4.03PVPTASHH219 pKa = 7.08DD220 pKa = 4.03APDD223 pKa = 4.33GGAPCDD229 pKa = 3.65PNSPQKK235 pKa = 10.99AKK237 pKa = 11.01APFSGKK243 pKa = 9.6RR244 pKa = 11.84KK245 pKa = 9.07HH246 pKa = 6.36NGPSVHH252 pKa = 6.4GSEE255 pKa = 5.13ASGPLLGGQGEE266 pKa = 4.62RR267 pKa = 11.84GPSPKK272 pKa = 9.39RR273 pKa = 11.84HH274 pKa = 5.97KK275 pKa = 9.8PASDD279 pKa = 3.59GEE281 pKa = 4.38RR282 pKa = 11.84TLRR285 pKa = 11.84QPTHH289 pKa = 6.42AAHH292 pKa = 6.47PTGTPVSLHH301 pKa = 5.51VQQQQQQHH309 pKa = 4.81QQQQLQQQQLQQLQQQQLQQQQQRR333 pKa = 11.84RR334 pKa = 11.84PQEE337 pKa = 3.98AEE339 pKa = 4.14DD340 pKa = 3.51RR341 pKa = 11.84HH342 pKa = 5.47QPRR345 pKa = 11.84ADD347 pKa = 3.39LPRR350 pKa = 11.84RR351 pKa = 11.84DD352 pKa = 4.31PLHH355 pKa = 6.42PASTPVPVIVLSGSPNQLKK374 pKa = 10.37CYY376 pKa = 10.18RR377 pKa = 11.84YY378 pKa = 10.07RR379 pKa = 11.84LNKK382 pKa = 9.26KK383 pKa = 9.04HH384 pKa = 6.21KK385 pKa = 9.63CLIKK389 pKa = 10.39FVSTTWYY396 pKa = 5.63WTNEE400 pKa = 3.47GGQGEE405 pKa = 4.5KK406 pKa = 11.23AMITLMFYY414 pKa = 10.85SEE416 pKa = 4.16ACKK419 pKa = 9.66NTFLNSVVLPKK430 pKa = 10.81GVTSTGGFLSLL441 pKa = 4.64
Molecular weight: 49.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2196 |
141 |
618 |
439.2 |
48.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.239 ± 0.538 | 2.322 ± 0.702 |
5.556 ± 0.544 | 5.191 ± 0.704 |
3.962 ± 0.421 | 7.24 ± 0.843 |
2.322 ± 0.259 | 3.506 ± 0.799 |
5.829 ± 0.962 | 8.106 ± 0.864 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.095 ± 0.286 | 4.053 ± 0.765 |
6.421 ± 1.116 | 5.055 ± 1.125 |
5.282 ± 0.161 | 8.333 ± 0.932 |
6.557 ± 1.02 | 6.922 ± 0.472 |
1.503 ± 0.379 | 3.506 ± 0.364 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
