
Hipposideros bat coronavirus HKU10
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Decacovirus; Bat coronavirus HKU10
Average proteome isoelectric point is 7.24
Get precalculated fractions of proteins
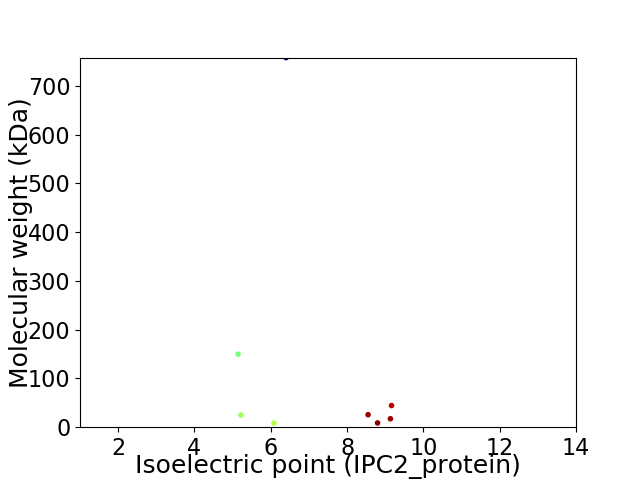
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
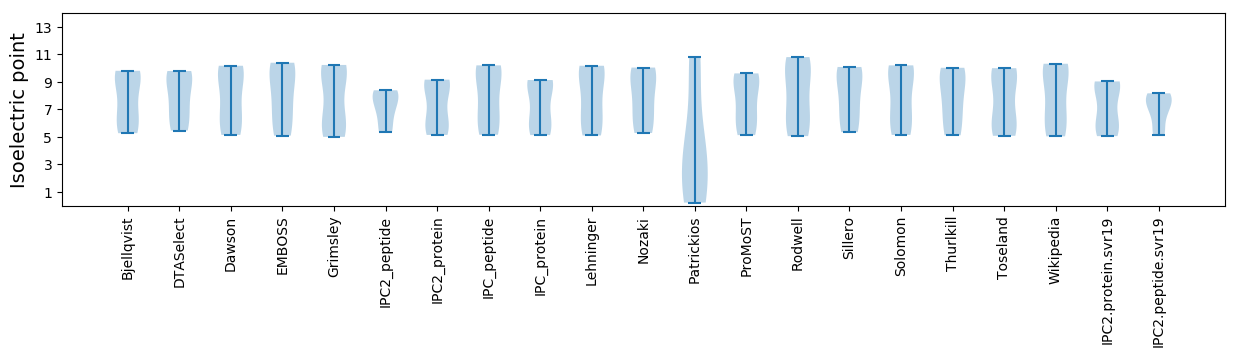
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K4JZR1|K4JZR1_9ALPC Envelope small membrane protein OS=Hipposideros bat coronavirus HKU10 OX=1241932 GN=E PE=3 SV=1
MM1 pKa = 7.54ISLAVYY7 pKa = 9.88LAEE10 pKa = 4.44LLVLITNLPYY20 pKa = 10.39TRR22 pKa = 11.84ATCSSAQNPAALPNLNLGLPANSNVFVSGYY52 pKa = 10.83LPIEE56 pKa = 4.32NNWRR60 pKa = 11.84CQTGDD65 pKa = 3.18TVYY68 pKa = 11.22NNVNAVYY75 pKa = 9.78WSYY78 pKa = 11.22YY79 pKa = 8.12SHH81 pKa = 6.45GTPVEE86 pKa = 3.99LAISNSDD93 pKa = 3.3QLQNADD99 pKa = 2.79QWALYY104 pKa = 9.59IYY106 pKa = 10.15QGNNYY111 pKa = 9.97GYY113 pKa = 8.51MHH115 pKa = 7.11FKK117 pKa = 8.59ICKK120 pKa = 7.96WNGKK124 pKa = 9.28YY125 pKa = 10.66SFINNPYY132 pKa = 8.81PSRR135 pKa = 11.84GVCMYY140 pKa = 10.3NKK142 pKa = 9.77KK143 pKa = 10.35FPFVFPHH150 pKa = 6.35ARR152 pKa = 11.84NTVIGASWSGDD163 pKa = 3.29YY164 pKa = 9.35VTLFTTTSARR174 pKa = 11.84RR175 pKa = 11.84FYY177 pKa = 11.26LKK179 pKa = 10.91NNWSRR184 pKa = 11.84VNVKK188 pKa = 10.12CQLAEE193 pKa = 3.84SCAIQTVSNINTLNITTNQQGFIQSYY219 pKa = 7.57SVCTEE224 pKa = 3.44CDD226 pKa = 3.29GFPKK230 pKa = 10.36HH231 pKa = 5.93VFPVLDD237 pKa = 4.46DD238 pKa = 4.02GKK240 pKa = 10.78IPGDD244 pKa = 3.37FSFSNWFVLTNSSTVLQGRR263 pKa = 11.84IVTVQPLKK271 pKa = 10.72LLCLWPVPSLNANDD285 pKa = 3.57KK286 pKa = 10.18PVYY289 pKa = 10.41FNLSLNSDD297 pKa = 3.68VRR299 pKa = 11.84CNGHH303 pKa = 6.53SIAGYY308 pKa = 9.05AQALRR313 pKa = 11.84FSLNFTANQVLNGVHH328 pKa = 6.46SIKK331 pKa = 10.63LFSSNQNFTFTCSTNGTLEE350 pKa = 4.59RR351 pKa = 11.84IPFGRR356 pKa = 11.84VEE358 pKa = 4.17SVYY361 pKa = 10.92KK362 pKa = 10.43CFVRR366 pKa = 11.84IDD368 pKa = 4.58AINSTKK374 pKa = 10.84NLFVGILPPVTKK386 pKa = 10.09EE387 pKa = 3.93FVISRR392 pKa = 11.84YY393 pKa = 8.85GAVYY397 pKa = 10.29INGVHH402 pKa = 6.55IMSLPPLEE410 pKa = 4.47SVIFNVTSSVGSDD423 pKa = 2.86FWTVAFADD431 pKa = 4.03EE432 pKa = 5.14AEE434 pKa = 4.28VLLDD438 pKa = 4.27LNATNIVDD446 pKa = 4.24ILYY449 pKa = 10.61CDD451 pKa = 3.59TLVNKK456 pKa = 10.05IKK458 pKa = 10.47CQNLAFSLEE467 pKa = 4.2DD468 pKa = 3.35GFYY471 pKa = 10.03ATSNLIEE478 pKa = 4.37RR479 pKa = 11.84DD480 pKa = 3.2IPRR483 pKa = 11.84SFITLPYY490 pKa = 9.48HH491 pKa = 4.73VTHH494 pKa = 6.33SVVGLDD500 pKa = 3.2VRR502 pKa = 11.84IYY504 pKa = 10.6YY505 pKa = 10.39EE506 pKa = 4.48NEE508 pKa = 3.48AASNLTLSPSDD519 pKa = 3.85PVCVDD524 pKa = 3.35SSQFSTYY531 pKa = 10.62FNVTADD537 pKa = 3.67TNGFNARR544 pKa = 11.84IINVDD549 pKa = 3.6CPFTFDD555 pKa = 4.45KK556 pKa = 10.85LNNYY560 pKa = 9.53ISFEE564 pKa = 4.42SICFSLEE571 pKa = 3.75PFAGGCSMRR580 pKa = 11.84IEE582 pKa = 4.73SFFSSYY588 pKa = 9.75VRR590 pKa = 11.84TVGSLYY596 pKa = 10.42ILFKK600 pKa = 10.84NGNKK604 pKa = 7.98ITGVTEE610 pKa = 4.26SSAGVYY616 pKa = 7.67DD617 pKa = 3.7TSFLHH622 pKa = 7.0TDD624 pKa = 2.39ICTDD628 pKa = 3.19YY629 pKa = 10.6TIYY632 pKa = 9.58GTTGRR637 pKa = 11.84GVITKK642 pKa = 10.54LNTTIIAGLYY652 pKa = 6.91YY653 pKa = 10.2TSQSGQILGFKK664 pKa = 10.15NATNGDD670 pKa = 3.56IYY672 pKa = 11.23AVRR675 pKa = 11.84PCDD678 pKa = 3.88LSSQAAVVSNQIVAVMSASEE698 pKa = 4.28NITFGFNNSLEE709 pKa = 4.12LPKK712 pKa = 10.48FYY714 pKa = 10.72LHH716 pKa = 7.09SNAEE720 pKa = 4.0NATNCTEE727 pKa = 3.78PVLIYY732 pKa = 10.46GKK734 pKa = 10.23IGICEE739 pKa = 4.43DD740 pKa = 3.64GSITEE745 pKa = 4.03VKK747 pKa = 10.08PRR749 pKa = 11.84IAEE752 pKa = 4.02PEE754 pKa = 4.27PYY756 pKa = 10.41SPIVTANITIPSNFSISVQAEE777 pKa = 4.07YY778 pKa = 11.36VQMFLTPVSVDD789 pKa = 2.89CNMYY793 pKa = 8.67VCNGNRR799 pKa = 11.84HH800 pKa = 5.67CLRR803 pKa = 11.84LLGEE807 pKa = 4.35YY808 pKa = 8.5VTACKK813 pKa = 10.52NIEE816 pKa = 3.94AALQLSARR824 pKa = 11.84LEE826 pKa = 4.39SIEE829 pKa = 4.0VNNFITVSEE838 pKa = 4.18NAISLANISNFDD850 pKa = 3.75SYY852 pKa = 11.7NLSVLLPKK860 pKa = 10.6QNGKK864 pKa = 9.43SVIEE868 pKa = 5.22DD869 pKa = 3.19ILFDD873 pKa = 3.77KK874 pKa = 11.17VVTNGLGTVDD884 pKa = 3.88QDD886 pKa = 3.64YY887 pKa = 10.93KK888 pKa = 11.0EE889 pKa = 4.41CIKK892 pKa = 10.77RR893 pKa = 11.84SGVKK897 pKa = 10.13DD898 pKa = 3.69VADD901 pKa = 3.83VACAQYY907 pKa = 11.33YY908 pKa = 9.05NGIMVLPGVVDD919 pKa = 3.87DD920 pKa = 5.03VKK922 pKa = 10.74MGLYY926 pKa = 8.32TASLTGAMVMGGLTAAAAIPFSIAVQSRR954 pKa = 11.84LNYY957 pKa = 9.76VALQTDD963 pKa = 4.37VLQKK967 pKa = 10.4NQQILADD974 pKa = 3.9SFNNAMSNITMAFAQVNDD992 pKa = 4.76AIKK995 pKa = 9.04EE996 pKa = 4.23TSQAISTVAQALSKK1010 pKa = 9.8VQSVVNEE1017 pKa = 3.85QGEE1020 pKa = 4.4ALSQLTKK1027 pKa = 10.68QLASNFQAISSSIEE1041 pKa = 4.09DD1042 pKa = 3.27IYY1044 pKa = 11.77NRR1046 pKa = 11.84LDD1048 pKa = 3.58MIAADD1053 pKa = 3.79AQVDD1057 pKa = 3.76RR1058 pKa = 11.84LITGRR1063 pKa = 11.84IGALNAFVTQTLTKK1077 pKa = 8.05YY1078 pKa = 9.0TEE1080 pKa = 4.13VRR1082 pKa = 11.84ASRR1085 pKa = 11.84QLALQKK1091 pKa = 10.44INEE1094 pKa = 4.49CVKK1097 pKa = 10.17SQSSRR1102 pKa = 11.84FGFCGNGTHH1111 pKa = 7.24LFSIANAAPNGIMLFHH1127 pKa = 6.46TVLMPTEE1134 pKa = 4.21YY1135 pKa = 10.84VVVRR1139 pKa = 11.84AWAGLCIEE1147 pKa = 5.14DD1148 pKa = 3.77KK1149 pKa = 11.3AFVLRR1154 pKa = 11.84DD1155 pKa = 3.42VKK1157 pKa = 9.04TTLFKK1162 pKa = 10.76TGDD1165 pKa = 3.21TYY1167 pKa = 11.66YY1168 pKa = 9.45ITTRR1172 pKa = 11.84DD1173 pKa = 3.53MYY1175 pKa = 10.55QPRR1178 pKa = 11.84LPQVSDD1184 pKa = 3.59FVQIKK1189 pKa = 9.92DD1190 pKa = 3.85CQVDD1194 pKa = 3.89YY1195 pKa = 11.75LNLTSDD1201 pKa = 3.74EE1202 pKa = 4.43VNNVIPDD1209 pKa = 3.89YY1210 pKa = 11.16IDD1212 pKa = 3.32VNKK1215 pKa = 9.54TLEE1218 pKa = 4.33DD1219 pKa = 3.97FGQILANNTRR1229 pKa = 11.84PPIPDD1234 pKa = 3.71FGLDD1238 pKa = 3.41VYY1240 pKa = 11.78NNTILNLTAEE1250 pKa = 4.39IDD1252 pKa = 3.73VLQNKK1257 pKa = 9.45SEE1259 pKa = 4.0QLLASTEE1266 pKa = 4.02RR1267 pKa = 11.84LQQLIDD1273 pKa = 4.05NLNKK1277 pKa = 10.23TYY1279 pKa = 11.22VDD1281 pKa = 4.26LEE1283 pKa = 3.94WLNRR1287 pKa = 11.84FEE1289 pKa = 6.35QYY1291 pKa = 10.44IKK1293 pKa = 9.88WPWYY1297 pKa = 7.76VWLAIFLAVILFSFLMLYY1315 pKa = 9.17CCCATGCCGCFSCLSNSCDD1334 pKa = 3.54CRR1336 pKa = 11.84GKK1338 pKa = 9.87NLQRR1342 pKa = 11.84YY1343 pKa = 6.8EE1344 pKa = 4.04VEE1346 pKa = 4.22KK1347 pKa = 11.1VHH1349 pKa = 6.52IQQ1351 pKa = 3.09
MM1 pKa = 7.54ISLAVYY7 pKa = 9.88LAEE10 pKa = 4.44LLVLITNLPYY20 pKa = 10.39TRR22 pKa = 11.84ATCSSAQNPAALPNLNLGLPANSNVFVSGYY52 pKa = 10.83LPIEE56 pKa = 4.32NNWRR60 pKa = 11.84CQTGDD65 pKa = 3.18TVYY68 pKa = 11.22NNVNAVYY75 pKa = 9.78WSYY78 pKa = 11.22YY79 pKa = 8.12SHH81 pKa = 6.45GTPVEE86 pKa = 3.99LAISNSDD93 pKa = 3.3QLQNADD99 pKa = 2.79QWALYY104 pKa = 9.59IYY106 pKa = 10.15QGNNYY111 pKa = 9.97GYY113 pKa = 8.51MHH115 pKa = 7.11FKK117 pKa = 8.59ICKK120 pKa = 7.96WNGKK124 pKa = 9.28YY125 pKa = 10.66SFINNPYY132 pKa = 8.81PSRR135 pKa = 11.84GVCMYY140 pKa = 10.3NKK142 pKa = 9.77KK143 pKa = 10.35FPFVFPHH150 pKa = 6.35ARR152 pKa = 11.84NTVIGASWSGDD163 pKa = 3.29YY164 pKa = 9.35VTLFTTTSARR174 pKa = 11.84RR175 pKa = 11.84FYY177 pKa = 11.26LKK179 pKa = 10.91NNWSRR184 pKa = 11.84VNVKK188 pKa = 10.12CQLAEE193 pKa = 3.84SCAIQTVSNINTLNITTNQQGFIQSYY219 pKa = 7.57SVCTEE224 pKa = 3.44CDD226 pKa = 3.29GFPKK230 pKa = 10.36HH231 pKa = 5.93VFPVLDD237 pKa = 4.46DD238 pKa = 4.02GKK240 pKa = 10.78IPGDD244 pKa = 3.37FSFSNWFVLTNSSTVLQGRR263 pKa = 11.84IVTVQPLKK271 pKa = 10.72LLCLWPVPSLNANDD285 pKa = 3.57KK286 pKa = 10.18PVYY289 pKa = 10.41FNLSLNSDD297 pKa = 3.68VRR299 pKa = 11.84CNGHH303 pKa = 6.53SIAGYY308 pKa = 9.05AQALRR313 pKa = 11.84FSLNFTANQVLNGVHH328 pKa = 6.46SIKK331 pKa = 10.63LFSSNQNFTFTCSTNGTLEE350 pKa = 4.59RR351 pKa = 11.84IPFGRR356 pKa = 11.84VEE358 pKa = 4.17SVYY361 pKa = 10.92KK362 pKa = 10.43CFVRR366 pKa = 11.84IDD368 pKa = 4.58AINSTKK374 pKa = 10.84NLFVGILPPVTKK386 pKa = 10.09EE387 pKa = 3.93FVISRR392 pKa = 11.84YY393 pKa = 8.85GAVYY397 pKa = 10.29INGVHH402 pKa = 6.55IMSLPPLEE410 pKa = 4.47SVIFNVTSSVGSDD423 pKa = 2.86FWTVAFADD431 pKa = 4.03EE432 pKa = 5.14AEE434 pKa = 4.28VLLDD438 pKa = 4.27LNATNIVDD446 pKa = 4.24ILYY449 pKa = 10.61CDD451 pKa = 3.59TLVNKK456 pKa = 10.05IKK458 pKa = 10.47CQNLAFSLEE467 pKa = 4.2DD468 pKa = 3.35GFYY471 pKa = 10.03ATSNLIEE478 pKa = 4.37RR479 pKa = 11.84DD480 pKa = 3.2IPRR483 pKa = 11.84SFITLPYY490 pKa = 9.48HH491 pKa = 4.73VTHH494 pKa = 6.33SVVGLDD500 pKa = 3.2VRR502 pKa = 11.84IYY504 pKa = 10.6YY505 pKa = 10.39EE506 pKa = 4.48NEE508 pKa = 3.48AASNLTLSPSDD519 pKa = 3.85PVCVDD524 pKa = 3.35SSQFSTYY531 pKa = 10.62FNVTADD537 pKa = 3.67TNGFNARR544 pKa = 11.84IINVDD549 pKa = 3.6CPFTFDD555 pKa = 4.45KK556 pKa = 10.85LNNYY560 pKa = 9.53ISFEE564 pKa = 4.42SICFSLEE571 pKa = 3.75PFAGGCSMRR580 pKa = 11.84IEE582 pKa = 4.73SFFSSYY588 pKa = 9.75VRR590 pKa = 11.84TVGSLYY596 pKa = 10.42ILFKK600 pKa = 10.84NGNKK604 pKa = 7.98ITGVTEE610 pKa = 4.26SSAGVYY616 pKa = 7.67DD617 pKa = 3.7TSFLHH622 pKa = 7.0TDD624 pKa = 2.39ICTDD628 pKa = 3.19YY629 pKa = 10.6TIYY632 pKa = 9.58GTTGRR637 pKa = 11.84GVITKK642 pKa = 10.54LNTTIIAGLYY652 pKa = 6.91YY653 pKa = 10.2TSQSGQILGFKK664 pKa = 10.15NATNGDD670 pKa = 3.56IYY672 pKa = 11.23AVRR675 pKa = 11.84PCDD678 pKa = 3.88LSSQAAVVSNQIVAVMSASEE698 pKa = 4.28NITFGFNNSLEE709 pKa = 4.12LPKK712 pKa = 10.48FYY714 pKa = 10.72LHH716 pKa = 7.09SNAEE720 pKa = 4.0NATNCTEE727 pKa = 3.78PVLIYY732 pKa = 10.46GKK734 pKa = 10.23IGICEE739 pKa = 4.43DD740 pKa = 3.64GSITEE745 pKa = 4.03VKK747 pKa = 10.08PRR749 pKa = 11.84IAEE752 pKa = 4.02PEE754 pKa = 4.27PYY756 pKa = 10.41SPIVTANITIPSNFSISVQAEE777 pKa = 4.07YY778 pKa = 11.36VQMFLTPVSVDD789 pKa = 2.89CNMYY793 pKa = 8.67VCNGNRR799 pKa = 11.84HH800 pKa = 5.67CLRR803 pKa = 11.84LLGEE807 pKa = 4.35YY808 pKa = 8.5VTACKK813 pKa = 10.52NIEE816 pKa = 3.94AALQLSARR824 pKa = 11.84LEE826 pKa = 4.39SIEE829 pKa = 4.0VNNFITVSEE838 pKa = 4.18NAISLANISNFDD850 pKa = 3.75SYY852 pKa = 11.7NLSVLLPKK860 pKa = 10.6QNGKK864 pKa = 9.43SVIEE868 pKa = 5.22DD869 pKa = 3.19ILFDD873 pKa = 3.77KK874 pKa = 11.17VVTNGLGTVDD884 pKa = 3.88QDD886 pKa = 3.64YY887 pKa = 10.93KK888 pKa = 11.0EE889 pKa = 4.41CIKK892 pKa = 10.77RR893 pKa = 11.84SGVKK897 pKa = 10.13DD898 pKa = 3.69VADD901 pKa = 3.83VACAQYY907 pKa = 11.33YY908 pKa = 9.05NGIMVLPGVVDD919 pKa = 3.87DD920 pKa = 5.03VKK922 pKa = 10.74MGLYY926 pKa = 8.32TASLTGAMVMGGLTAAAAIPFSIAVQSRR954 pKa = 11.84LNYY957 pKa = 9.76VALQTDD963 pKa = 4.37VLQKK967 pKa = 10.4NQQILADD974 pKa = 3.9SFNNAMSNITMAFAQVNDD992 pKa = 4.76AIKK995 pKa = 9.04EE996 pKa = 4.23TSQAISTVAQALSKK1010 pKa = 9.8VQSVVNEE1017 pKa = 3.85QGEE1020 pKa = 4.4ALSQLTKK1027 pKa = 10.68QLASNFQAISSSIEE1041 pKa = 4.09DD1042 pKa = 3.27IYY1044 pKa = 11.77NRR1046 pKa = 11.84LDD1048 pKa = 3.58MIAADD1053 pKa = 3.79AQVDD1057 pKa = 3.76RR1058 pKa = 11.84LITGRR1063 pKa = 11.84IGALNAFVTQTLTKK1077 pKa = 8.05YY1078 pKa = 9.0TEE1080 pKa = 4.13VRR1082 pKa = 11.84ASRR1085 pKa = 11.84QLALQKK1091 pKa = 10.44INEE1094 pKa = 4.49CVKK1097 pKa = 10.17SQSSRR1102 pKa = 11.84FGFCGNGTHH1111 pKa = 7.24LFSIANAAPNGIMLFHH1127 pKa = 6.46TVLMPTEE1134 pKa = 4.21YY1135 pKa = 10.84VVVRR1139 pKa = 11.84AWAGLCIEE1147 pKa = 5.14DD1148 pKa = 3.77KK1149 pKa = 11.3AFVLRR1154 pKa = 11.84DD1155 pKa = 3.42VKK1157 pKa = 9.04TTLFKK1162 pKa = 10.76TGDD1165 pKa = 3.21TYY1167 pKa = 11.66YY1168 pKa = 9.45ITTRR1172 pKa = 11.84DD1173 pKa = 3.53MYY1175 pKa = 10.55QPRR1178 pKa = 11.84LPQVSDD1184 pKa = 3.59FVQIKK1189 pKa = 9.92DD1190 pKa = 3.85CQVDD1194 pKa = 3.89YY1195 pKa = 11.75LNLTSDD1201 pKa = 3.74EE1202 pKa = 4.43VNNVIPDD1209 pKa = 3.89YY1210 pKa = 11.16IDD1212 pKa = 3.32VNKK1215 pKa = 9.54TLEE1218 pKa = 4.33DD1219 pKa = 3.97FGQILANNTRR1229 pKa = 11.84PPIPDD1234 pKa = 3.71FGLDD1238 pKa = 3.41VYY1240 pKa = 11.78NNTILNLTAEE1250 pKa = 4.39IDD1252 pKa = 3.73VLQNKK1257 pKa = 9.45SEE1259 pKa = 4.0QLLASTEE1266 pKa = 4.02RR1267 pKa = 11.84LQQLIDD1273 pKa = 4.05NLNKK1277 pKa = 10.23TYY1279 pKa = 11.22VDD1281 pKa = 4.26LEE1283 pKa = 3.94WLNRR1287 pKa = 11.84FEE1289 pKa = 6.35QYY1291 pKa = 10.44IKK1293 pKa = 9.88WPWYY1297 pKa = 7.76VWLAIFLAVILFSFLMLYY1315 pKa = 9.17CCCATGCCGCFSCLSNSCDD1334 pKa = 3.54CRR1336 pKa = 11.84GKK1338 pKa = 9.87NLQRR1342 pKa = 11.84YY1343 pKa = 6.8EE1344 pKa = 4.04VEE1346 pKa = 4.22KK1347 pKa = 11.1VHH1349 pKa = 6.52IQQ1351 pKa = 3.09
Molecular weight: 149.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4K1W8|K4K1W8_9ALPC Non-structural protein NS7b OS=Hipposideros bat coronavirus HKU10 OX=1241932 GN=NS7b PE=4 SV=1
MM1 pKa = 7.72ASGGNNVSFANKK13 pKa = 8.8PRR15 pKa = 11.84GRR17 pKa = 11.84SGRR20 pKa = 11.84VPLSYY25 pKa = 10.44YY26 pKa = 10.07SPVMIDD32 pKa = 3.05GDD34 pKa = 3.97QPFWKK39 pKa = 10.36VMPTNAVPTGMGDD52 pKa = 3.06KK53 pKa = 10.15NQQIGYY59 pKa = 9.11WNEE62 pKa = 3.52QPRR65 pKa = 11.84WRR67 pKa = 11.84MVKK70 pKa = 9.7GQRR73 pKa = 11.84KK74 pKa = 7.82EE75 pKa = 4.33LQSKK79 pKa = 6.13WHH81 pKa = 6.6FYY83 pKa = 11.09YY84 pKa = 10.85LGTGPQEE91 pKa = 4.08DD92 pKa = 3.12ARR94 pKa = 11.84YY95 pKa = 7.83RR96 pKa = 11.84QRR98 pKa = 11.84IEE100 pKa = 3.74GVFWVAVQGSKK111 pKa = 9.59TDD113 pKa = 3.49PTGLGTRR120 pKa = 11.84RR121 pKa = 11.84KK122 pKa = 8.71GQNLISPKK130 pKa = 9.89FAVKK134 pKa = 10.13IPSNVVVVEE143 pKa = 4.29EE144 pKa = 4.29TSRR147 pKa = 11.84PPSRR151 pKa = 11.84SQSNSRR157 pKa = 11.84PQSRR161 pKa = 11.84NNSRR165 pKa = 11.84PQSKK169 pKa = 10.58NNSQNNSRR177 pKa = 11.84DD178 pKa = 3.29NSRR181 pKa = 11.84APSRR185 pKa = 11.84PRR187 pKa = 11.84SRR189 pKa = 11.84ANSTSSSANDD199 pKa = 3.39AVDD202 pKa = 3.31IVAAVKK208 pKa = 10.1QALKK212 pKa = 10.26EE213 pKa = 4.02LGVTPEE219 pKa = 4.56KK220 pKa = 10.79KK221 pKa = 10.09NQEE224 pKa = 4.04KK225 pKa = 10.12QKK227 pKa = 10.76KK228 pKa = 8.81SNSGSNTPKK237 pKa = 10.02EE238 pKa = 4.25AATSRR243 pKa = 11.84QKK245 pKa = 10.91SSPSTPRR252 pKa = 11.84KK253 pKa = 8.69QLEE256 pKa = 3.86RR257 pKa = 11.84PEE259 pKa = 4.19WKK261 pKa = 9.81RR262 pKa = 11.84VPNKK266 pKa = 9.77EE267 pKa = 4.14EE268 pKa = 3.79NVIACFGPRR277 pKa = 11.84DD278 pKa = 3.77TLHH281 pKa = 6.7NFGDD285 pKa = 3.51QKK287 pKa = 11.22LIAEE291 pKa = 5.32GVQAPHH297 pKa = 6.22YY298 pKa = 7.82PQLAEE303 pKa = 4.7LVPTPAALLFGGEE316 pKa = 3.86VSTRR320 pKa = 11.84EE321 pKa = 3.78RR322 pKa = 11.84GDD324 pKa = 3.41EE325 pKa = 4.01VEE327 pKa = 4.28IMYY330 pKa = 9.8VYY332 pKa = 10.7KK333 pKa = 10.53MKK335 pKa = 10.38VHH337 pKa = 7.05KK338 pKa = 10.43SNKK341 pKa = 9.17DD342 pKa = 3.3LPSFLQQVSAYY353 pKa = 9.58AQAPDD358 pKa = 3.13ATTTEE363 pKa = 4.34KK364 pKa = 10.66ATVQPLLNPTAPDD377 pKa = 4.32FKK379 pKa = 10.45PATSDD384 pKa = 3.57EE385 pKa = 4.53VIEE388 pKa = 4.78MINTVYY394 pKa = 11.1DD395 pKa = 3.54SFDD398 pKa = 3.03AA399 pKa = 4.89
MM1 pKa = 7.72ASGGNNVSFANKK13 pKa = 8.8PRR15 pKa = 11.84GRR17 pKa = 11.84SGRR20 pKa = 11.84VPLSYY25 pKa = 10.44YY26 pKa = 10.07SPVMIDD32 pKa = 3.05GDD34 pKa = 3.97QPFWKK39 pKa = 10.36VMPTNAVPTGMGDD52 pKa = 3.06KK53 pKa = 10.15NQQIGYY59 pKa = 9.11WNEE62 pKa = 3.52QPRR65 pKa = 11.84WRR67 pKa = 11.84MVKK70 pKa = 9.7GQRR73 pKa = 11.84KK74 pKa = 7.82EE75 pKa = 4.33LQSKK79 pKa = 6.13WHH81 pKa = 6.6FYY83 pKa = 11.09YY84 pKa = 10.85LGTGPQEE91 pKa = 4.08DD92 pKa = 3.12ARR94 pKa = 11.84YY95 pKa = 7.83RR96 pKa = 11.84QRR98 pKa = 11.84IEE100 pKa = 3.74GVFWVAVQGSKK111 pKa = 9.59TDD113 pKa = 3.49PTGLGTRR120 pKa = 11.84RR121 pKa = 11.84KK122 pKa = 8.71GQNLISPKK130 pKa = 9.89FAVKK134 pKa = 10.13IPSNVVVVEE143 pKa = 4.29EE144 pKa = 4.29TSRR147 pKa = 11.84PPSRR151 pKa = 11.84SQSNSRR157 pKa = 11.84PQSRR161 pKa = 11.84NNSRR165 pKa = 11.84PQSKK169 pKa = 10.58NNSQNNSRR177 pKa = 11.84DD178 pKa = 3.29NSRR181 pKa = 11.84APSRR185 pKa = 11.84PRR187 pKa = 11.84SRR189 pKa = 11.84ANSTSSSANDD199 pKa = 3.39AVDD202 pKa = 3.31IVAAVKK208 pKa = 10.1QALKK212 pKa = 10.26EE213 pKa = 4.02LGVTPEE219 pKa = 4.56KK220 pKa = 10.79KK221 pKa = 10.09NQEE224 pKa = 4.04KK225 pKa = 10.12QKK227 pKa = 10.76KK228 pKa = 8.81SNSGSNTPKK237 pKa = 10.02EE238 pKa = 4.25AATSRR243 pKa = 11.84QKK245 pKa = 10.91SSPSTPRR252 pKa = 11.84KK253 pKa = 8.69QLEE256 pKa = 3.86RR257 pKa = 11.84PEE259 pKa = 4.19WKK261 pKa = 9.81RR262 pKa = 11.84VPNKK266 pKa = 9.77EE267 pKa = 4.14EE268 pKa = 3.79NVIACFGPRR277 pKa = 11.84DD278 pKa = 3.77TLHH281 pKa = 6.7NFGDD285 pKa = 3.51QKK287 pKa = 11.22LIAEE291 pKa = 5.32GVQAPHH297 pKa = 6.22YY298 pKa = 7.82PQLAEE303 pKa = 4.7LVPTPAALLFGGEE316 pKa = 3.86VSTRR320 pKa = 11.84EE321 pKa = 3.78RR322 pKa = 11.84GDD324 pKa = 3.41EE325 pKa = 4.01VEE327 pKa = 4.28IMYY330 pKa = 9.8VYY332 pKa = 10.7KK333 pKa = 10.53MKK335 pKa = 10.38VHH337 pKa = 7.05KK338 pKa = 10.43SNKK341 pKa = 9.17DD342 pKa = 3.3LPSFLQQVSAYY353 pKa = 9.58AQAPDD358 pKa = 3.13ATTTEE363 pKa = 4.34KK364 pKa = 10.66ATVQPLLNPTAPDD377 pKa = 4.32FKK379 pKa = 10.45PATSDD384 pKa = 3.57EE385 pKa = 4.53VIEE388 pKa = 4.78MINTVYY394 pKa = 11.1DD395 pKa = 3.54SFDD398 pKa = 3.03AA399 pKa = 4.89
Molecular weight: 44.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9279 |
75 |
6781 |
1159.9 |
129.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.466 ± 0.254 | 3.222 ± 0.721 |
5.151 ± 0.575 | 4.171 ± 0.233 |
5.582 ± 0.432 | 6.1 ± 0.585 |
1.875 ± 0.286 | 5.216 ± 0.559 |
5.787 ± 0.848 | 8.816 ± 0.567 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.145 ± 0.249 | 5.884 ± 0.766 |
3.675 ± 0.691 | 3.567 ± 0.565 |
3.449 ± 0.51 | 7.167 ± 0.616 |
6.24 ± 0.405 | 9.613 ± 0.493 |
1.25 ± 0.27 | 4.623 ± 0.279 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
