
Dankookia rubra
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Dankookia
Average proteome isoelectric point is 7.13
Get precalculated fractions of proteins
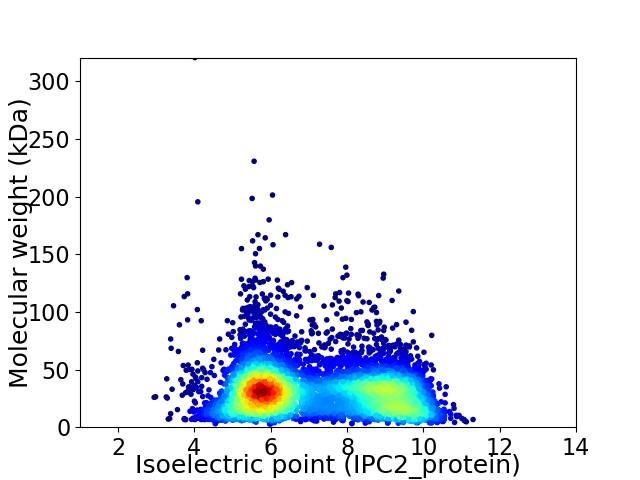
Virtual 2D-PAGE plot for 7087 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
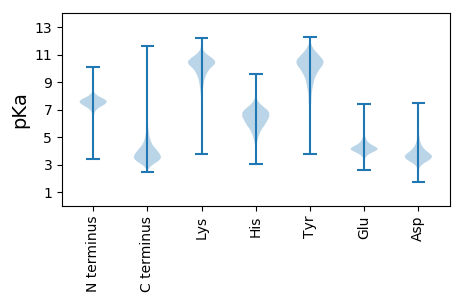
Protein with the lowest isoelectric point:
>tr|A0A4V3AA02|A0A4V3AA02_9PROT IS21 family transposase OS=Dankookia rubra OX=1442381 GN=E2C06_17690 PE=4 SV=1
MM1 pKa = 7.71SGTTSNTAFGADD13 pKa = 3.96DD14 pKa = 3.78SAIPAAIGGGGSAGPSVGAGDD35 pKa = 3.6ATGPIVSVNTSGIDD49 pKa = 3.66LAGLPAQYY57 pKa = 9.26STASADD63 pKa = 3.57GTGLGFFTRR72 pKa = 11.84QLEE75 pKa = 4.32YY76 pKa = 10.97VPTAVTVGPDD86 pKa = 3.14DD87 pKa = 3.92ALYY90 pKa = 10.89VSGLSGLPYY99 pKa = 9.16PDD101 pKa = 4.12GYY103 pKa = 10.61SRR105 pKa = 11.84VMRR108 pKa = 11.84VADD111 pKa = 3.6THH113 pKa = 5.97ATTGFDD119 pKa = 3.56GQIDD123 pKa = 3.73SGVPQTYY130 pKa = 10.6ASGLSQVNGLSFDD143 pKa = 3.69GKK145 pKa = 11.2GNLNVLEE152 pKa = 4.55FVNAANIYY160 pKa = 10.62DD161 pKa = 4.1PTRR164 pKa = 11.84QPGEE168 pKa = 4.23LPPSEE173 pKa = 5.59LIRR176 pKa = 11.84IAPDD180 pKa = 3.55GVRR183 pKa = 11.84TTISGPEE190 pKa = 3.95LKK192 pKa = 10.43LANYY196 pKa = 9.37VLADD200 pKa = 3.67KK201 pKa = 8.78EE202 pKa = 4.5TGDD205 pKa = 3.69VYY207 pKa = 11.37VAIGNADD214 pKa = 3.36INNGEE219 pKa = 4.29VLRR222 pKa = 11.84YY223 pKa = 9.58HH224 pKa = 7.09VDD226 pKa = 3.45DD227 pKa = 4.09ATGQATSVDD236 pKa = 4.09VVASGLNNPRR246 pKa = 11.84GMAFGPDD253 pKa = 2.51GHH255 pKa = 8.37LYY257 pKa = 10.97VNEE260 pKa = 4.22QGLGTPGDD268 pKa = 4.04SPDD271 pKa = 3.54AATAPTIPFIPGAVDD286 pKa = 3.12EE287 pKa = 5.15RR288 pKa = 11.84GGFTASITRR297 pKa = 11.84VDD299 pKa = 3.24IDD301 pKa = 3.43GSGQEE306 pKa = 4.65RR307 pKa = 11.84IFTGLPSIRR316 pKa = 11.84EE317 pKa = 3.98VNAATGEE324 pKa = 4.0DD325 pKa = 3.52RR326 pKa = 11.84VISVGSNGFTIAPDD340 pKa = 3.47GTAYY344 pKa = 10.86LATGGGLTAATADD357 pKa = 3.76ALGPLAPYY365 pKa = 9.96IQGLLKK371 pKa = 9.9IDD373 pKa = 3.84GLFDD377 pKa = 4.64HH378 pKa = 7.52DD379 pKa = 4.47PSNVTVTPAFNSVTYY394 pKa = 10.23AAEE397 pKa = 4.23NGPDD401 pKa = 3.64GSQTLFNTQSNLNDD415 pKa = 3.33VVVGSDD421 pKa = 2.79GHH423 pKa = 7.93IYY425 pKa = 10.73AVDD428 pKa = 3.25AARR431 pKa = 11.84NVMYY435 pKa = 10.51GFSPDD440 pKa = 4.35DD441 pKa = 3.83LSTPDD446 pKa = 3.72SVTVFQKK453 pKa = 10.17QAPVLTPPQYY463 pKa = 10.86AAVVAAGGNPSLDD476 pKa = 3.65YY477 pKa = 10.56QVEE480 pKa = 4.33IAQTTYY486 pKa = 10.64KK487 pKa = 10.6AEE489 pKa = 4.28NGAPDD494 pKa = 3.7TPGRR498 pKa = 11.84AEE500 pKa = 3.84AAASFSPADD509 pKa = 3.97PGAAAAGAPAGPADD523 pKa = 4.02AGLPAGIVDD532 pKa = 3.72TAVPPRR538 pKa = 11.84GEE540 pKa = 4.19DD541 pKa = 3.27ASVGTVNAAGADD553 pKa = 3.93GLPGSIPVTDD563 pKa = 4.28PSFPGPIDD571 pKa = 4.69PISPPVLPGNVYY583 pKa = 10.09VPYY586 pKa = 10.13FDD588 pKa = 4.79PFFGGNYY595 pKa = 10.01APATPPVLPAGGHH608 pKa = 5.46GATYY612 pKa = 9.22TVSHH616 pKa = 7.13LYY618 pKa = 10.8SFGDD622 pKa = 3.49RR623 pKa = 11.84LVDD626 pKa = 4.28DD627 pKa = 4.7GGSYY631 pKa = 10.14GAAAVAQAAGQPAPNASPVYY651 pKa = 8.49YY652 pKa = 10.25QGGFSDD658 pKa = 4.41GPNWTDD664 pKa = 3.13NLAKK668 pKa = 10.36ILGAQQTDD676 pKa = 3.85GEE678 pKa = 4.9DD679 pKa = 3.35NNFGYY684 pKa = 10.85VLATARR690 pKa = 11.84PIEE693 pKa = 4.4NPLDD697 pKa = 3.99PFTGQTTLNTFEE709 pKa = 4.18GQIDD713 pKa = 4.19AFQQAHH719 pKa = 6.82AYY721 pKa = 9.94FSANDD726 pKa = 3.72LVTVTFGGNDD736 pKa = 3.17LTLPSNLSPEE746 pKa = 4.16EE747 pKa = 4.74GITEE751 pKa = 4.19SVQAIVDD758 pKa = 3.86GLDD761 pKa = 2.97RR762 pKa = 11.84LADD765 pKa = 4.15LGAHH769 pKa = 6.19HH770 pKa = 6.94FVITNLPDD778 pKa = 3.91LEE780 pKa = 4.37LAPLFKK786 pKa = 11.06DD787 pKa = 3.65PDD789 pKa = 3.74FLQQLGAEE797 pKa = 4.48PGSFQPLVAEE807 pKa = 5.05FNARR811 pKa = 11.84LADD814 pKa = 3.61AVHH817 pKa = 6.39AFEE820 pKa = 4.97GEE822 pKa = 4.0RR823 pKa = 11.84GVDD826 pKa = 3.38VTLLDD831 pKa = 3.51VHH833 pKa = 7.21KK834 pKa = 10.57LFDD837 pKa = 6.01AIADD841 pKa = 3.77DD842 pKa = 3.87PAAYY846 pKa = 10.03GFNDD850 pKa = 2.86VDD852 pKa = 4.11QPVLAAPPLQPGTPTVYY869 pKa = 10.41NPAIVGQDD877 pKa = 3.36PAVEE881 pKa = 4.05HH882 pKa = 6.88ASLFIDD888 pKa = 4.14PFFHH892 pKa = 6.61PTALGQAILAEE903 pKa = 4.52TAWHH907 pKa = 6.4VIAA910 pKa = 5.91
MM1 pKa = 7.71SGTTSNTAFGADD13 pKa = 3.96DD14 pKa = 3.78SAIPAAIGGGGSAGPSVGAGDD35 pKa = 3.6ATGPIVSVNTSGIDD49 pKa = 3.66LAGLPAQYY57 pKa = 9.26STASADD63 pKa = 3.57GTGLGFFTRR72 pKa = 11.84QLEE75 pKa = 4.32YY76 pKa = 10.97VPTAVTVGPDD86 pKa = 3.14DD87 pKa = 3.92ALYY90 pKa = 10.89VSGLSGLPYY99 pKa = 9.16PDD101 pKa = 4.12GYY103 pKa = 10.61SRR105 pKa = 11.84VMRR108 pKa = 11.84VADD111 pKa = 3.6THH113 pKa = 5.97ATTGFDD119 pKa = 3.56GQIDD123 pKa = 3.73SGVPQTYY130 pKa = 10.6ASGLSQVNGLSFDD143 pKa = 3.69GKK145 pKa = 11.2GNLNVLEE152 pKa = 4.55FVNAANIYY160 pKa = 10.62DD161 pKa = 4.1PTRR164 pKa = 11.84QPGEE168 pKa = 4.23LPPSEE173 pKa = 5.59LIRR176 pKa = 11.84IAPDD180 pKa = 3.55GVRR183 pKa = 11.84TTISGPEE190 pKa = 3.95LKK192 pKa = 10.43LANYY196 pKa = 9.37VLADD200 pKa = 3.67KK201 pKa = 8.78EE202 pKa = 4.5TGDD205 pKa = 3.69VYY207 pKa = 11.37VAIGNADD214 pKa = 3.36INNGEE219 pKa = 4.29VLRR222 pKa = 11.84YY223 pKa = 9.58HH224 pKa = 7.09VDD226 pKa = 3.45DD227 pKa = 4.09ATGQATSVDD236 pKa = 4.09VVASGLNNPRR246 pKa = 11.84GMAFGPDD253 pKa = 2.51GHH255 pKa = 8.37LYY257 pKa = 10.97VNEE260 pKa = 4.22QGLGTPGDD268 pKa = 4.04SPDD271 pKa = 3.54AATAPTIPFIPGAVDD286 pKa = 3.12EE287 pKa = 5.15RR288 pKa = 11.84GGFTASITRR297 pKa = 11.84VDD299 pKa = 3.24IDD301 pKa = 3.43GSGQEE306 pKa = 4.65RR307 pKa = 11.84IFTGLPSIRR316 pKa = 11.84EE317 pKa = 3.98VNAATGEE324 pKa = 4.0DD325 pKa = 3.52RR326 pKa = 11.84VISVGSNGFTIAPDD340 pKa = 3.47GTAYY344 pKa = 10.86LATGGGLTAATADD357 pKa = 3.76ALGPLAPYY365 pKa = 9.96IQGLLKK371 pKa = 9.9IDD373 pKa = 3.84GLFDD377 pKa = 4.64HH378 pKa = 7.52DD379 pKa = 4.47PSNVTVTPAFNSVTYY394 pKa = 10.23AAEE397 pKa = 4.23NGPDD401 pKa = 3.64GSQTLFNTQSNLNDD415 pKa = 3.33VVVGSDD421 pKa = 2.79GHH423 pKa = 7.93IYY425 pKa = 10.73AVDD428 pKa = 3.25AARR431 pKa = 11.84NVMYY435 pKa = 10.51GFSPDD440 pKa = 4.35DD441 pKa = 3.83LSTPDD446 pKa = 3.72SVTVFQKK453 pKa = 10.17QAPVLTPPQYY463 pKa = 10.86AAVVAAGGNPSLDD476 pKa = 3.65YY477 pKa = 10.56QVEE480 pKa = 4.33IAQTTYY486 pKa = 10.64KK487 pKa = 10.6AEE489 pKa = 4.28NGAPDD494 pKa = 3.7TPGRR498 pKa = 11.84AEE500 pKa = 3.84AAASFSPADD509 pKa = 3.97PGAAAAGAPAGPADD523 pKa = 4.02AGLPAGIVDD532 pKa = 3.72TAVPPRR538 pKa = 11.84GEE540 pKa = 4.19DD541 pKa = 3.27ASVGTVNAAGADD553 pKa = 3.93GLPGSIPVTDD563 pKa = 4.28PSFPGPIDD571 pKa = 4.69PISPPVLPGNVYY583 pKa = 10.09VPYY586 pKa = 10.13FDD588 pKa = 4.79PFFGGNYY595 pKa = 10.01APATPPVLPAGGHH608 pKa = 5.46GATYY612 pKa = 9.22TVSHH616 pKa = 7.13LYY618 pKa = 10.8SFGDD622 pKa = 3.49RR623 pKa = 11.84LVDD626 pKa = 4.28DD627 pKa = 4.7GGSYY631 pKa = 10.14GAAAVAQAAGQPAPNASPVYY651 pKa = 8.49YY652 pKa = 10.25QGGFSDD658 pKa = 4.41GPNWTDD664 pKa = 3.13NLAKK668 pKa = 10.36ILGAQQTDD676 pKa = 3.85GEE678 pKa = 4.9DD679 pKa = 3.35NNFGYY684 pKa = 10.85VLATARR690 pKa = 11.84PIEE693 pKa = 4.4NPLDD697 pKa = 3.99PFTGQTTLNTFEE709 pKa = 4.18GQIDD713 pKa = 4.19AFQQAHH719 pKa = 6.82AYY721 pKa = 9.94FSANDD726 pKa = 3.72LVTVTFGGNDD736 pKa = 3.17LTLPSNLSPEE746 pKa = 4.16EE747 pKa = 4.74GITEE751 pKa = 4.19SVQAIVDD758 pKa = 3.86GLDD761 pKa = 2.97RR762 pKa = 11.84LADD765 pKa = 4.15LGAHH769 pKa = 6.19HH770 pKa = 6.94FVITNLPDD778 pKa = 3.91LEE780 pKa = 4.37LAPLFKK786 pKa = 11.06DD787 pKa = 3.65PDD789 pKa = 3.74FLQQLGAEE797 pKa = 4.48PGSFQPLVAEE807 pKa = 5.05FNARR811 pKa = 11.84LADD814 pKa = 3.61AVHH817 pKa = 6.39AFEE820 pKa = 4.97GEE822 pKa = 4.0RR823 pKa = 11.84GVDD826 pKa = 3.38VTLLDD831 pKa = 3.51VHH833 pKa = 7.21KK834 pKa = 10.57LFDD837 pKa = 6.01AIADD841 pKa = 3.77DD842 pKa = 3.87PAAYY846 pKa = 10.03GFNDD850 pKa = 2.86VDD852 pKa = 4.11QPVLAAPPLQPGTPTVYY869 pKa = 10.41NPAIVGQDD877 pKa = 3.36PAVEE881 pKa = 4.05HH882 pKa = 6.88ASLFIDD888 pKa = 4.14PFFHH892 pKa = 6.61PTALGQAILAEE903 pKa = 4.52TAWHH907 pKa = 6.4VIAA910 pKa = 5.91
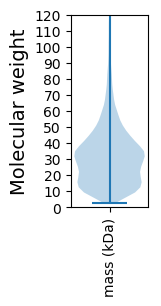
Molecular weight: 93.09 kDa
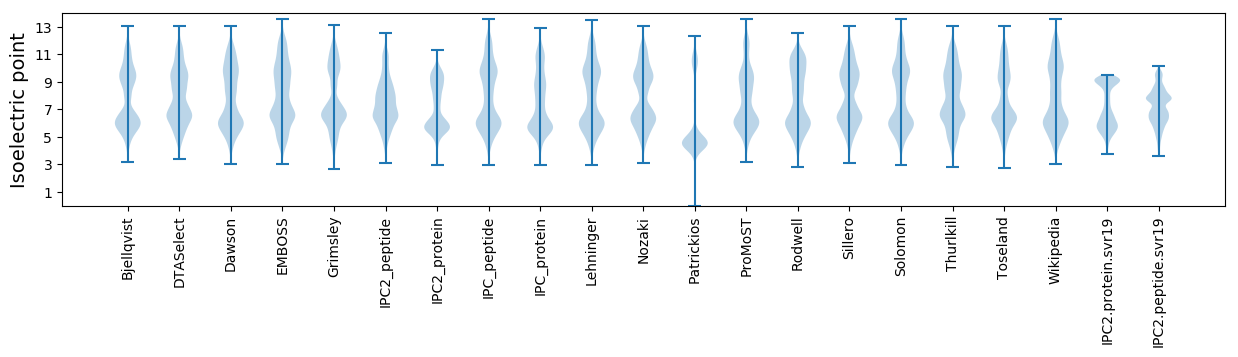
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R5Q7N2|A0A4R5Q7N2_9PROT PaaI family thioesterase OS=Dankookia rubra OX=1442381 GN=E2C06_32165 PE=4 SV=1
MM1 pKa = 7.59PPAPARR7 pKa = 11.84AAGRR11 pKa = 11.84APAAAPRR18 pKa = 11.84GRR20 pKa = 11.84ARR22 pKa = 11.84GGGRR26 pKa = 11.84TAPARR31 pKa = 11.84PSAPPAPAPAWSRR44 pKa = 11.84RR45 pKa = 11.84APRR48 pKa = 11.84PAGPAPPLAPPGPAPGRR65 pKa = 11.84RR66 pKa = 11.84PRR68 pKa = 3.85
MM1 pKa = 7.59PPAPARR7 pKa = 11.84AAGRR11 pKa = 11.84APAAAPRR18 pKa = 11.84GRR20 pKa = 11.84ARR22 pKa = 11.84GGGRR26 pKa = 11.84TAPARR31 pKa = 11.84PSAPPAPAPAWSRR44 pKa = 11.84RR45 pKa = 11.84APRR48 pKa = 11.84PAGPAPPLAPPGPAPGRR65 pKa = 11.84RR66 pKa = 11.84PRR68 pKa = 3.85
Molecular weight: 6.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2193505 |
25 |
3143 |
309.5 |
33.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.024 ± 0.05 | 0.908 ± 0.01 |
5.042 ± 0.022 | 5.335 ± 0.025 |
3.211 ± 0.016 | 9.512 ± 0.029 |
2.047 ± 0.014 | 3.893 ± 0.018 |
2.023 ± 0.022 | 10.911 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.294 ± 0.014 | 1.984 ± 0.015 |
6.364 ± 0.025 | 3.064 ± 0.018 |
8.445 ± 0.033 | 4.332 ± 0.019 |
4.944 ± 0.02 | 7.31 ± 0.026 |
1.567 ± 0.013 | 1.79 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
