
Avon-Heathcote Estuary associated circular virus 16
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.75
Get precalculated fractions of proteins
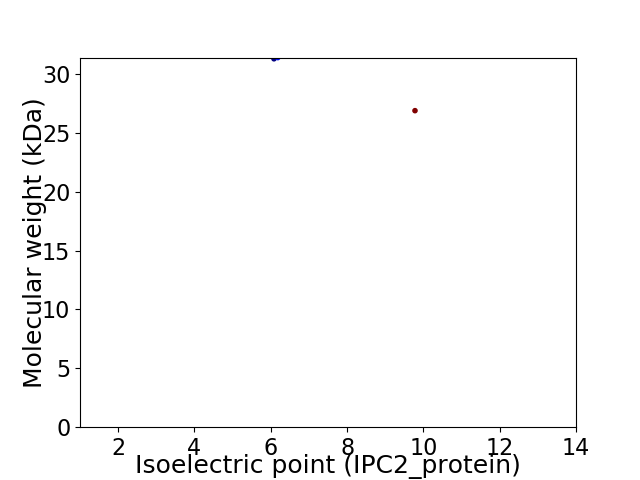
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IBQ8|A0A0C5IBQ8_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 16 OX=1618239 PE=3 SV=1
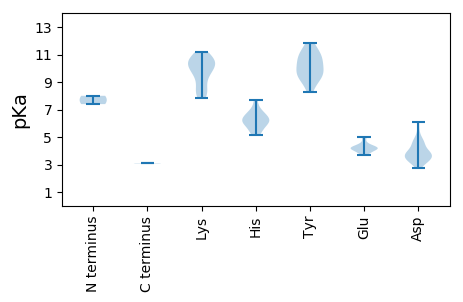
MM1 pKa = 8.0ADD3 pKa = 3.67RR4 pKa = 11.84RR5 pKa = 11.84PHH7 pKa = 6.61KK8 pKa = 10.5SWTYY12 pKa = 9.61TLNEE16 pKa = 4.14PTDD19 pKa = 3.59EE20 pKa = 4.48DD21 pKa = 3.92EE22 pKa = 5.02NRR24 pKa = 11.84IHH26 pKa = 6.7LWDD29 pKa = 3.71VSRR32 pKa = 11.84QIYY35 pKa = 8.89GHH37 pKa = 5.54EE38 pKa = 4.09TAPTTGQHH46 pKa = 5.97HH47 pKa = 6.25LQGFVSFRR55 pKa = 11.84AAKK58 pKa = 10.09RR59 pKa = 11.84FTGVTRR65 pKa = 11.84LLPRR69 pKa = 11.84AHH71 pKa = 6.78WEE73 pKa = 3.91VAKK76 pKa = 10.64YY77 pKa = 9.78PSSAWDD83 pKa = 3.55YY84 pKa = 8.3CTKK87 pKa = 10.87GDD89 pKa = 4.61DD90 pKa = 4.76LYY92 pKa = 11.87VVDD95 pKa = 4.21NRR97 pKa = 11.84VGRR100 pKa = 11.84GTRR103 pKa = 11.84SDD105 pKa = 3.32WQDD108 pKa = 3.13VLDD111 pKa = 4.61LVSEE115 pKa = 4.53GASDD119 pKa = 3.9AAILRR124 pKa = 11.84AQPSCFFKK132 pKa = 11.09YY133 pKa = 10.28GAGISRR139 pKa = 11.84ARR141 pKa = 11.84AALQEE146 pKa = 4.22PRR148 pKa = 11.84HH149 pKa = 5.78HH150 pKa = 5.89QTLCEE155 pKa = 4.13WYY157 pKa = 9.76YY158 pKa = 11.43GPTGGGKK165 pKa = 7.87STYY168 pKa = 9.42IAQEE172 pKa = 3.82HH173 pKa = 7.05PEE175 pKa = 4.26ADD177 pKa = 2.78WCQITKK183 pKa = 10.5SGFLLGYY190 pKa = 9.13HH191 pKa = 6.46NKK193 pKa = 8.0PTVVFDD199 pKa = 6.06DD200 pKa = 4.55PDD202 pKa = 4.6LDD204 pKa = 3.76AFGRR208 pKa = 11.84EE209 pKa = 3.91LFLNLVNRR217 pKa = 11.84TPFEE221 pKa = 4.29MNVKK225 pKa = 8.31GTSVAFNAKK234 pKa = 9.8LVIVASNKK242 pKa = 9.96HH243 pKa = 6.21PDD245 pKa = 3.38CFIPGDD251 pKa = 3.7AAVHH255 pKa = 5.31RR256 pKa = 11.84RR257 pKa = 11.84IHH259 pKa = 5.17KK260 pKa = 8.68TFLVSCIDD268 pKa = 3.6GEE270 pKa = 4.91HH271 pKa = 6.12MVEE274 pKa = 4.22QQQ276 pKa = 3.08
MM1 pKa = 8.0ADD3 pKa = 3.67RR4 pKa = 11.84RR5 pKa = 11.84PHH7 pKa = 6.61KK8 pKa = 10.5SWTYY12 pKa = 9.61TLNEE16 pKa = 4.14PTDD19 pKa = 3.59EE20 pKa = 4.48DD21 pKa = 3.92EE22 pKa = 5.02NRR24 pKa = 11.84IHH26 pKa = 6.7LWDD29 pKa = 3.71VSRR32 pKa = 11.84QIYY35 pKa = 8.89GHH37 pKa = 5.54EE38 pKa = 4.09TAPTTGQHH46 pKa = 5.97HH47 pKa = 6.25LQGFVSFRR55 pKa = 11.84AAKK58 pKa = 10.09RR59 pKa = 11.84FTGVTRR65 pKa = 11.84LLPRR69 pKa = 11.84AHH71 pKa = 6.78WEE73 pKa = 3.91VAKK76 pKa = 10.64YY77 pKa = 9.78PSSAWDD83 pKa = 3.55YY84 pKa = 8.3CTKK87 pKa = 10.87GDD89 pKa = 4.61DD90 pKa = 4.76LYY92 pKa = 11.87VVDD95 pKa = 4.21NRR97 pKa = 11.84VGRR100 pKa = 11.84GTRR103 pKa = 11.84SDD105 pKa = 3.32WQDD108 pKa = 3.13VLDD111 pKa = 4.61LVSEE115 pKa = 4.53GASDD119 pKa = 3.9AAILRR124 pKa = 11.84AQPSCFFKK132 pKa = 11.09YY133 pKa = 10.28GAGISRR139 pKa = 11.84ARR141 pKa = 11.84AALQEE146 pKa = 4.22PRR148 pKa = 11.84HH149 pKa = 5.78HH150 pKa = 5.89QTLCEE155 pKa = 4.13WYY157 pKa = 9.76YY158 pKa = 11.43GPTGGGKK165 pKa = 7.87STYY168 pKa = 9.42IAQEE172 pKa = 3.82HH173 pKa = 7.05PEE175 pKa = 4.26ADD177 pKa = 2.78WCQITKK183 pKa = 10.5SGFLLGYY190 pKa = 9.13HH191 pKa = 6.46NKK193 pKa = 8.0PTVVFDD199 pKa = 6.06DD200 pKa = 4.55PDD202 pKa = 4.6LDD204 pKa = 3.76AFGRR208 pKa = 11.84EE209 pKa = 3.91LFLNLVNRR217 pKa = 11.84TPFEE221 pKa = 4.29MNVKK225 pKa = 8.31GTSVAFNAKK234 pKa = 9.8LVIVASNKK242 pKa = 9.96HH243 pKa = 6.21PDD245 pKa = 3.38CFIPGDD251 pKa = 3.7AAVHH255 pKa = 5.31RR256 pKa = 11.84RR257 pKa = 11.84IHH259 pKa = 5.17KK260 pKa = 8.68TFLVSCIDD268 pKa = 3.6GEE270 pKa = 4.91HH271 pKa = 6.12MVEE274 pKa = 4.22QQQ276 pKa = 3.08
Molecular weight: 31.28 kDa
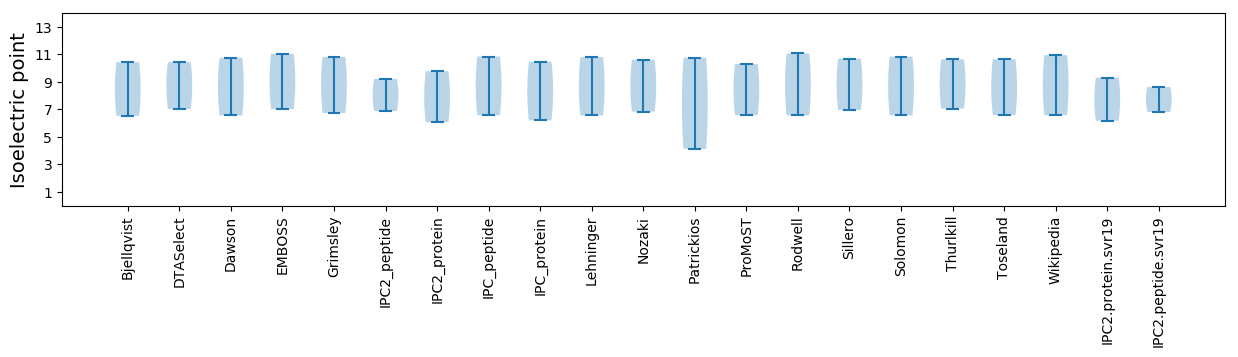
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IBQ8|A0A0C5IBQ8_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 16 OX=1618239 PE=3 SV=1
MM1 pKa = 7.39ARR3 pKa = 11.84ATVRR7 pKa = 11.84QGNVALAKK15 pKa = 9.65RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84ARR21 pKa = 11.84GRR23 pKa = 11.84KK24 pKa = 8.24IRR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 9.44GRR30 pKa = 11.84RR31 pKa = 11.84KK32 pKa = 10.04AITARR37 pKa = 11.84RR38 pKa = 11.84VKK40 pKa = 10.68AIVSKK45 pKa = 10.67SISKK49 pKa = 10.28RR50 pKa = 11.84VEE52 pKa = 3.71TKK54 pKa = 9.14TQVIDD59 pKa = 3.27VTTASGGTWYY69 pKa = 10.65GFTINEE75 pKa = 4.13IPQGDD80 pKa = 3.83TSITRR85 pKa = 11.84DD86 pKa = 3.38GTSVFMKK93 pKa = 10.39SLQLNIMNVMSATTPTGTAYY113 pKa = 10.66DD114 pKa = 3.77YY115 pKa = 11.15MRR117 pKa = 11.84FILVNFPDD125 pKa = 4.87GEE127 pKa = 4.27QGDD130 pKa = 4.11ITDD133 pKa = 3.87VLAIDD138 pKa = 4.19TAPTSASALTMPYY151 pKa = 10.37RR152 pKa = 11.84MKK154 pKa = 10.68RR155 pKa = 11.84DD156 pKa = 3.56TVGVATHH163 pKa = 6.22TIKK166 pKa = 11.15YY167 pKa = 9.32QVLADD172 pKa = 3.25WHH174 pKa = 6.26EE175 pKa = 4.22LLNEE179 pKa = 4.06STHH182 pKa = 7.66AIDD185 pKa = 5.36LNRR188 pKa = 11.84LRR190 pKa = 11.84LKK192 pKa = 10.44INKK195 pKa = 9.03KK196 pKa = 8.22CTYY199 pKa = 11.05ALGNTDD205 pKa = 3.11GSVITKK211 pKa = 9.88NRR213 pKa = 11.84IRR215 pKa = 11.84LFYY218 pKa = 10.97YY219 pKa = 9.72GLQSIAEE226 pKa = 4.29TGTNIKK232 pKa = 9.99GRR234 pKa = 11.84LYY236 pKa = 10.26FTDD239 pKa = 3.25SS240 pKa = 3.11
MM1 pKa = 7.39ARR3 pKa = 11.84ATVRR7 pKa = 11.84QGNVALAKK15 pKa = 9.65RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84ARR21 pKa = 11.84GRR23 pKa = 11.84KK24 pKa = 8.24IRR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 9.44GRR30 pKa = 11.84RR31 pKa = 11.84KK32 pKa = 10.04AITARR37 pKa = 11.84RR38 pKa = 11.84VKK40 pKa = 10.68AIVSKK45 pKa = 10.67SISKK49 pKa = 10.28RR50 pKa = 11.84VEE52 pKa = 3.71TKK54 pKa = 9.14TQVIDD59 pKa = 3.27VTTASGGTWYY69 pKa = 10.65GFTINEE75 pKa = 4.13IPQGDD80 pKa = 3.83TSITRR85 pKa = 11.84DD86 pKa = 3.38GTSVFMKK93 pKa = 10.39SLQLNIMNVMSATTPTGTAYY113 pKa = 10.66DD114 pKa = 3.77YY115 pKa = 11.15MRR117 pKa = 11.84FILVNFPDD125 pKa = 4.87GEE127 pKa = 4.27QGDD130 pKa = 4.11ITDD133 pKa = 3.87VLAIDD138 pKa = 4.19TAPTSASALTMPYY151 pKa = 10.37RR152 pKa = 11.84MKK154 pKa = 10.68RR155 pKa = 11.84DD156 pKa = 3.56TVGVATHH163 pKa = 6.22TIKK166 pKa = 11.15YY167 pKa = 9.32QVLADD172 pKa = 3.25WHH174 pKa = 6.26EE175 pKa = 4.22LLNEE179 pKa = 4.06STHH182 pKa = 7.66AIDD185 pKa = 5.36LNRR188 pKa = 11.84LRR190 pKa = 11.84LKK192 pKa = 10.44INKK195 pKa = 9.03KK196 pKa = 8.22CTYY199 pKa = 11.05ALGNTDD205 pKa = 3.11GSVITKK211 pKa = 9.88NRR213 pKa = 11.84IRR215 pKa = 11.84LFYY218 pKa = 10.97YY219 pKa = 9.72GLQSIAEE226 pKa = 4.29TGTNIKK232 pKa = 9.99GRR234 pKa = 11.84LYY236 pKa = 10.26FTDD239 pKa = 3.25SS240 pKa = 3.11
Molecular weight: 26.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
516 |
240 |
276 |
258.0 |
29.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.333 ± 0.0 | 1.357 ± 0.629 |
6.395 ± 0.654 | 3.876 ± 0.92 |
3.682 ± 0.791 | 7.171 ± 0.058 |
3.295 ± 1.367 | 5.62 ± 1.536 |
5.426 ± 0.829 | 6.783 ± 0.078 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.938 ± 0.654 | 3.876 ± 0.473 |
3.682 ± 1.069 | 3.488 ± 0.382 |
8.333 ± 1.115 | 5.62 ± 0.143 |
9.109 ± 1.989 | 6.589 ± 0.227 |
1.744 ± 0.609 | 3.682 ± 0.045 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
