
Human papillomavirus 165
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 12
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
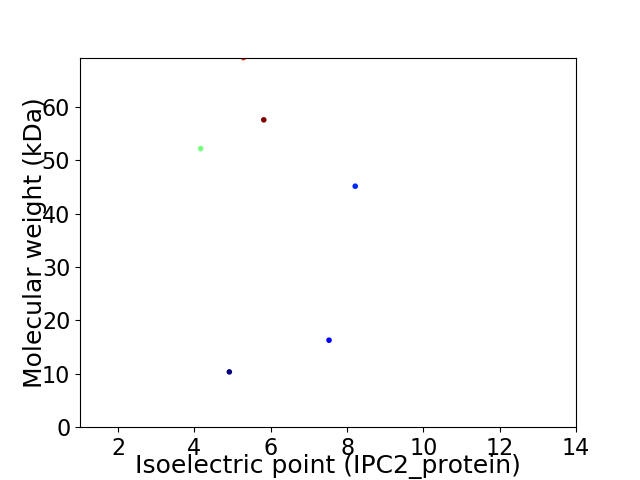
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
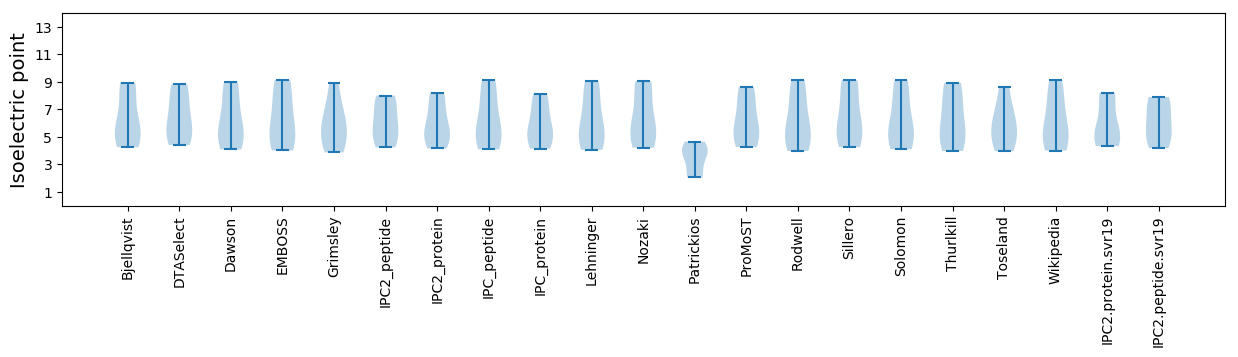
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K4MZ15|K4MZ15_9PAPI Minor capsid protein L2 OS=Human papillomavirus 165 OX=1315266 GN=L2 PE=3 SV=1
MM1 pKa = 8.1PDD3 pKa = 3.1VQNKK7 pKa = 9.25FEE9 pKa = 4.23QKK11 pKa = 7.68TWADD15 pKa = 3.5TLLKK19 pKa = 10.44IFGSLLYY26 pKa = 10.37FGNLGIGTGRR36 pKa = 11.84GSGGTLGYY44 pKa = 10.16RR45 pKa = 11.84PLGTPSAPRR54 pKa = 11.84ATEE57 pKa = 3.74VTPIRR62 pKa = 11.84PNLTIDD68 pKa = 3.46PVGPTDD74 pKa = 3.41ITVVDD79 pKa = 4.5AAAPSIVPLSEE90 pKa = 3.76GVPDD94 pKa = 3.92ISYY97 pKa = 9.92IAPDD101 pKa = 3.97AGPGAGAEE109 pKa = 4.23DD110 pKa = 3.78IEE112 pKa = 5.46LFTITNPSTDD122 pKa = 3.02VGGAAVTPTVISTEE136 pKa = 3.97EE137 pKa = 3.94GAVAILDD144 pKa = 3.78TQTIPEE150 pKa = 4.16RR151 pKa = 11.84PVQVFYY157 pKa = 11.39DD158 pKa = 4.32PAATTTHH165 pKa = 7.22EE166 pKa = 4.52INVFAAPIEE175 pKa = 4.23TSSNVNVFVDD185 pKa = 4.12SNVSGTIIGGEE196 pKa = 4.24DD197 pKa = 3.11IPLEE201 pKa = 3.97RR202 pKa = 11.84LNYY205 pKa = 9.95SEE207 pKa = 5.68FEE209 pKa = 4.05IEE211 pKa = 4.17EE212 pKa = 4.48TPLTSTPTRR221 pKa = 11.84RR222 pKa = 11.84LEE224 pKa = 3.95SAINRR229 pKa = 11.84TKK231 pKa = 10.5GFYY234 pKa = 10.08RR235 pKa = 11.84RR236 pKa = 11.84FIKK239 pKa = 9.93QVPVRR244 pKa = 11.84SFEE247 pKa = 4.52FVTQPSRR254 pKa = 11.84LVEE257 pKa = 4.26FQFEE261 pKa = 4.27NPAFDD266 pKa = 4.81ADD268 pKa = 3.69VTLEE272 pKa = 3.87FEE274 pKa = 4.95RR275 pKa = 11.84DD276 pKa = 3.35LADD279 pKa = 3.24ITAAPADD286 pKa = 3.88EE287 pKa = 4.58FQDD290 pKa = 3.54VIRR293 pKa = 11.84LHH295 pKa = 6.84RR296 pKa = 11.84PLFSSVEE303 pKa = 3.9GAVRR307 pKa = 11.84VSRR310 pKa = 11.84LGDD313 pKa = 3.03TGTIRR318 pKa = 11.84TRR320 pKa = 11.84SGTIIGQQVHH330 pKa = 7.1FYY332 pKa = 10.77HH333 pKa = 7.55DD334 pKa = 3.09ISDD337 pKa = 3.52IVTEE341 pKa = 4.46EE342 pKa = 4.32IEE344 pKa = 4.62LEE346 pKa = 4.37TIPTEE351 pKa = 4.26PEE353 pKa = 3.52TSIVDD358 pKa = 4.27DD359 pKa = 4.15IMDD362 pKa = 4.02SVIVDD367 pKa = 4.91PINTADD373 pKa = 3.51VAIAEE378 pKa = 4.15EE379 pKa = 4.76DD380 pKa = 3.73LLDD383 pKa = 4.21SYY385 pKa = 11.91GEE387 pKa = 4.24NFAQTHH393 pKa = 6.93LILGDD398 pKa = 3.58VEE400 pKa = 4.71LAGEE404 pKa = 4.29PEE406 pKa = 4.03ILIIPITAEE415 pKa = 3.91TASLNTFIPALVDD428 pKa = 3.4ASIIDD433 pKa = 3.99ASSTAYY439 pKa = 9.71PIAPILPVPEE449 pKa = 4.54PAPHH453 pKa = 5.9TVSDD457 pKa = 4.04INDD460 pKa = 3.74DD461 pKa = 3.97YY462 pKa = 11.89YY463 pKa = 11.32LYY465 pKa = 10.32PSLYY469 pKa = 9.01PKK471 pKa = 10.27RR472 pKa = 11.84KK473 pKa = 9.26RR474 pKa = 11.84RR475 pKa = 11.84RR476 pKa = 11.84LDD478 pKa = 3.22FFF480 pKa = 5.54
MM1 pKa = 8.1PDD3 pKa = 3.1VQNKK7 pKa = 9.25FEE9 pKa = 4.23QKK11 pKa = 7.68TWADD15 pKa = 3.5TLLKK19 pKa = 10.44IFGSLLYY26 pKa = 10.37FGNLGIGTGRR36 pKa = 11.84GSGGTLGYY44 pKa = 10.16RR45 pKa = 11.84PLGTPSAPRR54 pKa = 11.84ATEE57 pKa = 3.74VTPIRR62 pKa = 11.84PNLTIDD68 pKa = 3.46PVGPTDD74 pKa = 3.41ITVVDD79 pKa = 4.5AAAPSIVPLSEE90 pKa = 3.76GVPDD94 pKa = 3.92ISYY97 pKa = 9.92IAPDD101 pKa = 3.97AGPGAGAEE109 pKa = 4.23DD110 pKa = 3.78IEE112 pKa = 5.46LFTITNPSTDD122 pKa = 3.02VGGAAVTPTVISTEE136 pKa = 3.97EE137 pKa = 3.94GAVAILDD144 pKa = 3.78TQTIPEE150 pKa = 4.16RR151 pKa = 11.84PVQVFYY157 pKa = 11.39DD158 pKa = 4.32PAATTTHH165 pKa = 7.22EE166 pKa = 4.52INVFAAPIEE175 pKa = 4.23TSSNVNVFVDD185 pKa = 4.12SNVSGTIIGGEE196 pKa = 4.24DD197 pKa = 3.11IPLEE201 pKa = 3.97RR202 pKa = 11.84LNYY205 pKa = 9.95SEE207 pKa = 5.68FEE209 pKa = 4.05IEE211 pKa = 4.17EE212 pKa = 4.48TPLTSTPTRR221 pKa = 11.84RR222 pKa = 11.84LEE224 pKa = 3.95SAINRR229 pKa = 11.84TKK231 pKa = 10.5GFYY234 pKa = 10.08RR235 pKa = 11.84RR236 pKa = 11.84FIKK239 pKa = 9.93QVPVRR244 pKa = 11.84SFEE247 pKa = 4.52FVTQPSRR254 pKa = 11.84LVEE257 pKa = 4.26FQFEE261 pKa = 4.27NPAFDD266 pKa = 4.81ADD268 pKa = 3.69VTLEE272 pKa = 3.87FEE274 pKa = 4.95RR275 pKa = 11.84DD276 pKa = 3.35LADD279 pKa = 3.24ITAAPADD286 pKa = 3.88EE287 pKa = 4.58FQDD290 pKa = 3.54VIRR293 pKa = 11.84LHH295 pKa = 6.84RR296 pKa = 11.84PLFSSVEE303 pKa = 3.9GAVRR307 pKa = 11.84VSRR310 pKa = 11.84LGDD313 pKa = 3.03TGTIRR318 pKa = 11.84TRR320 pKa = 11.84SGTIIGQQVHH330 pKa = 7.1FYY332 pKa = 10.77HH333 pKa = 7.55DD334 pKa = 3.09ISDD337 pKa = 3.52IVTEE341 pKa = 4.46EE342 pKa = 4.32IEE344 pKa = 4.62LEE346 pKa = 4.37TIPTEE351 pKa = 4.26PEE353 pKa = 3.52TSIVDD358 pKa = 4.27DD359 pKa = 4.15IMDD362 pKa = 4.02SVIVDD367 pKa = 4.91PINTADD373 pKa = 3.51VAIAEE378 pKa = 4.15EE379 pKa = 4.76DD380 pKa = 3.73LLDD383 pKa = 4.21SYY385 pKa = 11.91GEE387 pKa = 4.24NFAQTHH393 pKa = 6.93LILGDD398 pKa = 3.58VEE400 pKa = 4.71LAGEE404 pKa = 4.29PEE406 pKa = 4.03ILIIPITAEE415 pKa = 3.91TASLNTFIPALVDD428 pKa = 3.4ASIIDD433 pKa = 3.99ASSTAYY439 pKa = 9.71PIAPILPVPEE449 pKa = 4.54PAPHH453 pKa = 5.9TVSDD457 pKa = 4.04INDD460 pKa = 3.74DD461 pKa = 3.97YY462 pKa = 11.89YY463 pKa = 11.32LYY465 pKa = 10.32PSLYY469 pKa = 9.01PKK471 pKa = 10.27RR472 pKa = 11.84KK473 pKa = 9.26RR474 pKa = 11.84RR475 pKa = 11.84RR476 pKa = 11.84LDD478 pKa = 3.22FFF480 pKa = 5.54
Molecular weight: 52.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4MNC5|K4MNC5_9PAPI Protein E6 OS=Human papillomavirus 165 OX=1315266 GN=E6 PE=3 SV=1
MM1 pKa = 7.37EE2 pKa = 4.95TPQTLADD9 pKa = 3.92RR10 pKa = 11.84FAAQQEE16 pKa = 4.34KK17 pKa = 10.7QMTLIEE23 pKa = 4.37NEE25 pKa = 4.21STQLTDD31 pKa = 5.35HH32 pKa = 6.55IEE34 pKa = 4.01YY35 pKa = 9.22WNSVRR40 pKa = 11.84LEE42 pKa = 4.11NVLGYY47 pKa = 8.03YY48 pKa = 9.63ARR50 pKa = 11.84KK51 pKa = 9.82EE52 pKa = 4.03GLTQLGMQPLPVLQVLEE69 pKa = 4.54YY70 pKa = 9.82KK71 pKa = 10.67AKK73 pKa = 10.21EE74 pKa = 4.08AIKK77 pKa = 10.21MSLLLNSLNKK87 pKa = 9.85SEE89 pKa = 4.33YY90 pKa = 10.57GKK92 pKa = 10.58EE93 pKa = 3.68PWTLAEE99 pKa = 4.12VSAEE103 pKa = 3.75IVNTNPKK110 pKa = 10.1NCFKK114 pKa = 10.75KK115 pKa = 10.65KK116 pKa = 9.44PFTVTVYY123 pKa = 10.64FDD125 pKa = 4.16NDD127 pKa = 3.42EE128 pKa = 4.38QNSFPYY134 pKa = 10.01TNWDD138 pKa = 3.44SIYY141 pKa = 10.25YY142 pKa = 9.94QDD144 pKa = 5.91DD145 pKa = 3.35NSVWHH150 pKa = 6.36KK151 pKa = 10.68VHH153 pKa = 6.8GKK155 pKa = 9.85VDD157 pKa = 3.61TNGLYY162 pKa = 10.22FQEE165 pKa = 4.02ITGDD169 pKa = 3.06IVYY172 pKa = 9.6FALFQPDD179 pKa = 3.46AEE181 pKa = 4.65RR182 pKa = 11.84YY183 pKa = 7.77GHH185 pKa = 6.04SGQWSVKK192 pKa = 9.72YY193 pKa = 10.76KK194 pKa = 9.61NTTLFTSVTSSTPGPSSTEE213 pKa = 3.61PASQRR218 pKa = 11.84RR219 pKa = 11.84ATTHH223 pKa = 5.13TVSKK227 pKa = 10.64QKK229 pKa = 9.52TPRR232 pKa = 11.84KK233 pKa = 9.14RR234 pKa = 11.84KK235 pKa = 9.82HH236 pKa = 5.37EE237 pKa = 4.17TDD239 pKa = 3.14EE240 pKa = 4.46DD241 pKa = 4.32TDD243 pKa = 4.36GEE245 pKa = 4.85SPSSTSSGFRR255 pKa = 11.84LRR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84RR260 pKa = 11.84GEE262 pKa = 3.9QGEE265 pKa = 4.56STSRR269 pKa = 11.84ASSSRR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84GGNGGAVSPEE286 pKa = 3.94EE287 pKa = 3.97VGSRR291 pKa = 11.84TRR293 pKa = 11.84TVPTQGLTRR302 pKa = 11.84LRR304 pKa = 11.84RR305 pKa = 11.84LQEE308 pKa = 3.82EE309 pKa = 4.16ARR311 pKa = 11.84DD312 pKa = 3.71PPIIILQGCPNTLKK326 pKa = 10.66CFRR329 pKa = 11.84NRR331 pKa = 11.84VSHH334 pKa = 5.99KK335 pKa = 9.99HH336 pKa = 5.26HH337 pKa = 6.69SLYY340 pKa = 10.67TDD342 pKa = 3.33ATTVFHH348 pKa = 6.83WVLANCQDD356 pKa = 3.55KK357 pKa = 11.2KK358 pKa = 10.87EE359 pKa = 4.2SARR362 pKa = 11.84MLIAFTDD369 pKa = 3.23NRR371 pKa = 11.84QRR373 pKa = 11.84DD374 pKa = 3.61AFLAHH379 pKa = 5.32VTLPKK384 pKa = 9.69GTNHH388 pKa = 6.48WFGSLDD394 pKa = 3.56TLL396 pKa = 4.76
MM1 pKa = 7.37EE2 pKa = 4.95TPQTLADD9 pKa = 3.92RR10 pKa = 11.84FAAQQEE16 pKa = 4.34KK17 pKa = 10.7QMTLIEE23 pKa = 4.37NEE25 pKa = 4.21STQLTDD31 pKa = 5.35HH32 pKa = 6.55IEE34 pKa = 4.01YY35 pKa = 9.22WNSVRR40 pKa = 11.84LEE42 pKa = 4.11NVLGYY47 pKa = 8.03YY48 pKa = 9.63ARR50 pKa = 11.84KK51 pKa = 9.82EE52 pKa = 4.03GLTQLGMQPLPVLQVLEE69 pKa = 4.54YY70 pKa = 9.82KK71 pKa = 10.67AKK73 pKa = 10.21EE74 pKa = 4.08AIKK77 pKa = 10.21MSLLLNSLNKK87 pKa = 9.85SEE89 pKa = 4.33YY90 pKa = 10.57GKK92 pKa = 10.58EE93 pKa = 3.68PWTLAEE99 pKa = 4.12VSAEE103 pKa = 3.75IVNTNPKK110 pKa = 10.1NCFKK114 pKa = 10.75KK115 pKa = 10.65KK116 pKa = 9.44PFTVTVYY123 pKa = 10.64FDD125 pKa = 4.16NDD127 pKa = 3.42EE128 pKa = 4.38QNSFPYY134 pKa = 10.01TNWDD138 pKa = 3.44SIYY141 pKa = 10.25YY142 pKa = 9.94QDD144 pKa = 5.91DD145 pKa = 3.35NSVWHH150 pKa = 6.36KK151 pKa = 10.68VHH153 pKa = 6.8GKK155 pKa = 9.85VDD157 pKa = 3.61TNGLYY162 pKa = 10.22FQEE165 pKa = 4.02ITGDD169 pKa = 3.06IVYY172 pKa = 9.6FALFQPDD179 pKa = 3.46AEE181 pKa = 4.65RR182 pKa = 11.84YY183 pKa = 7.77GHH185 pKa = 6.04SGQWSVKK192 pKa = 9.72YY193 pKa = 10.76KK194 pKa = 9.61NTTLFTSVTSSTPGPSSTEE213 pKa = 3.61PASQRR218 pKa = 11.84RR219 pKa = 11.84ATTHH223 pKa = 5.13TVSKK227 pKa = 10.64QKK229 pKa = 9.52TPRR232 pKa = 11.84KK233 pKa = 9.14RR234 pKa = 11.84KK235 pKa = 9.82HH236 pKa = 5.37EE237 pKa = 4.17TDD239 pKa = 3.14EE240 pKa = 4.46DD241 pKa = 4.32TDD243 pKa = 4.36GEE245 pKa = 4.85SPSSTSSGFRR255 pKa = 11.84LRR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84RR260 pKa = 11.84GEE262 pKa = 3.9QGEE265 pKa = 4.56STSRR269 pKa = 11.84ASSSRR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84GGNGGAVSPEE286 pKa = 3.94EE287 pKa = 3.97VGSRR291 pKa = 11.84TRR293 pKa = 11.84TVPTQGLTRR302 pKa = 11.84LRR304 pKa = 11.84RR305 pKa = 11.84LQEE308 pKa = 3.82EE309 pKa = 4.16ARR311 pKa = 11.84DD312 pKa = 3.71PPIIILQGCPNTLKK326 pKa = 10.66CFRR329 pKa = 11.84NRR331 pKa = 11.84VSHH334 pKa = 5.99KK335 pKa = 9.99HH336 pKa = 5.26HH337 pKa = 6.69SLYY340 pKa = 10.67TDD342 pKa = 3.33ATTVFHH348 pKa = 6.83WVLANCQDD356 pKa = 3.55KK357 pKa = 11.2KK358 pKa = 10.87EE359 pKa = 4.2SARR362 pKa = 11.84MLIAFTDD369 pKa = 3.23NRR371 pKa = 11.84QRR373 pKa = 11.84DD374 pKa = 3.61AFLAHH379 pKa = 5.32VTLPKK384 pKa = 9.69GTNHH388 pKa = 6.48WFGSLDD394 pKa = 3.56TLL396 pKa = 4.76
Molecular weight: 45.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2222 |
93 |
608 |
370.3 |
41.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.986 ± 0.468 | 2.295 ± 0.813 |
6.391 ± 0.467 | 6.391 ± 0.476 |
5.041 ± 0.385 | 5.221 ± 0.766 |
1.935 ± 0.291 | 5.401 ± 0.894 |
4.995 ± 0.754 | 8.506 ± 0.624 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.26 ± 0.274 | 5.491 ± 0.782 |
5.896 ± 0.868 | 3.735 ± 0.381 |
5.311 ± 0.579 | 7.111 ± 0.765 |
7.381 ± 0.711 | 6.841 ± 0.438 |
1.305 ± 0.245 | 3.51 ± 0.19 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
