
Capybara microvirus Cap3_SP_478
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.76
Get precalculated fractions of proteins
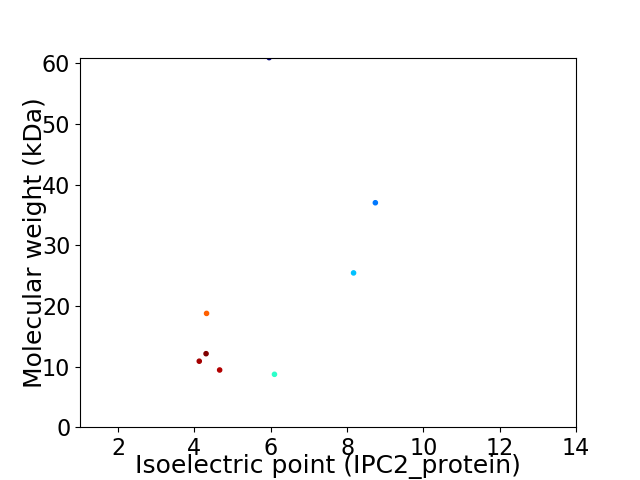
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
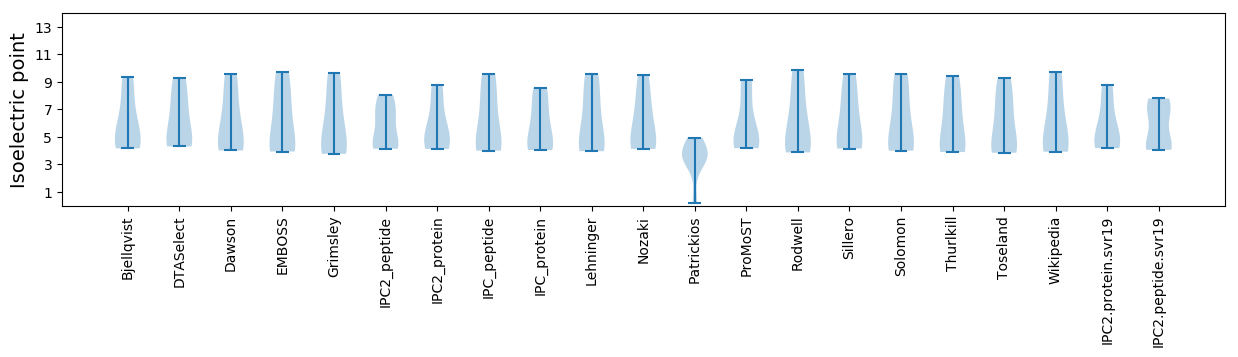
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W630|A0A4P8W630_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_478 OX=2585468 PE=4 SV=1
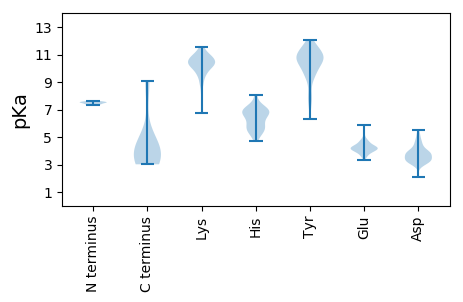
MM1 pKa = 7.62VNIKK5 pKa = 10.47YY6 pKa = 10.58SDD8 pKa = 3.27IAPILGEE15 pKa = 4.45HH16 pKa = 5.37YY17 pKa = 11.14LEE19 pKa = 4.75FKK21 pKa = 11.14FNGVWLKK28 pKa = 10.11TSILNNTDD36 pKa = 2.92SNNIVSIFMLNGEE49 pKa = 4.55DD50 pKa = 4.54LYY52 pKa = 11.69CEE54 pKa = 4.32VKK56 pKa = 10.59YY57 pKa = 10.74HH58 pKa = 6.98EE59 pKa = 4.61LLQYY63 pKa = 11.21VLVKK67 pKa = 9.58HH68 pKa = 6.25ICSDD72 pKa = 3.83YY73 pKa = 11.47YY74 pKa = 11.25DD75 pKa = 4.36IDD77 pKa = 3.05WDD79 pKa = 3.65YY80 pKa = 11.64SYY82 pKa = 11.92NEE84 pKa = 4.22VIEE87 pKa = 4.82CEE89 pKa = 4.32CEE91 pKa = 4.19VNFLNGEE98 pKa = 4.11EE99 pKa = 4.18NKK101 pKa = 10.29FF102 pKa = 3.5
MM1 pKa = 7.62VNIKK5 pKa = 10.47YY6 pKa = 10.58SDD8 pKa = 3.27IAPILGEE15 pKa = 4.45HH16 pKa = 5.37YY17 pKa = 11.14LEE19 pKa = 4.75FKK21 pKa = 11.14FNGVWLKK28 pKa = 10.11TSILNNTDD36 pKa = 2.92SNNIVSIFMLNGEE49 pKa = 4.55DD50 pKa = 4.54LYY52 pKa = 11.69CEE54 pKa = 4.32VKK56 pKa = 10.59YY57 pKa = 10.74HH58 pKa = 6.98EE59 pKa = 4.61LLQYY63 pKa = 11.21VLVKK67 pKa = 9.58HH68 pKa = 6.25ICSDD72 pKa = 3.83YY73 pKa = 11.47YY74 pKa = 11.25DD75 pKa = 4.36IDD77 pKa = 3.05WDD79 pKa = 3.65YY80 pKa = 11.64SYY82 pKa = 11.92NEE84 pKa = 4.22VIEE87 pKa = 4.82CEE89 pKa = 4.32CEE91 pKa = 4.19VNFLNGEE98 pKa = 4.11EE99 pKa = 4.18NKK101 pKa = 10.29FF102 pKa = 3.5
Molecular weight: 12.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W8R3|A0A4P8W8R3_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_478 OX=2585468 PE=3 SV=1
MM1 pKa = 7.56CVNPIKK7 pKa = 10.37CWQYY11 pKa = 11.4YY12 pKa = 6.65EE13 pKa = 4.34TDD15 pKa = 3.43SLTGEE20 pKa = 4.42EE21 pKa = 4.14KK22 pKa = 9.28THH24 pKa = 7.0LVFNINKK31 pKa = 8.99TSFNKK36 pKa = 10.63DD37 pKa = 2.08KK38 pKa = 10.85DD39 pKa = 3.86YY40 pKa = 11.54SVEE43 pKa = 4.07QKK45 pKa = 11.11LEE47 pKa = 3.84DD48 pKa = 3.84LYY50 pKa = 10.97IPGRR54 pKa = 11.84NQKK57 pKa = 10.18LYY59 pKa = 10.51NRR61 pKa = 11.84RR62 pKa = 11.84IKK64 pKa = 10.5PITLPCGKK72 pKa = 10.38CPDD75 pKa = 4.08CLMKK79 pKa = 10.94YY80 pKa = 9.4SLEE83 pKa = 3.64WAYY86 pKa = 11.05RR87 pKa = 11.84IMNEE91 pKa = 3.3ASYY94 pKa = 10.75YY95 pKa = 10.99AEE97 pKa = 4.13NCFMTLTYY105 pKa = 11.19ANTNGEE111 pKa = 4.37LCLKK115 pKa = 10.36DD116 pKa = 2.97IQNFLKK122 pKa = 10.63RR123 pKa = 11.84LRR125 pKa = 11.84KK126 pKa = 9.78KK127 pKa = 10.5ISPLKK132 pKa = 10.38IRR134 pKa = 11.84FFNCGEE140 pKa = 3.97YY141 pKa = 10.46GSKK144 pKa = 10.27GKK146 pKa = 10.24RR147 pKa = 11.84PHH149 pKa = 5.59FHH151 pKa = 7.07CIIFGWCPSDD161 pKa = 4.85LKK163 pKa = 10.97FLKK166 pKa = 9.02KK167 pKa = 8.76TKK169 pKa = 9.72KK170 pKa = 10.11GQIIYY175 pKa = 9.24TSEE178 pKa = 3.86IVSSLWKK185 pKa = 10.35HH186 pKa = 5.31GFVSIGNVEE195 pKa = 4.19FEE197 pKa = 4.41SAKK200 pKa = 9.4YY201 pKa = 7.37TALYY205 pKa = 7.84MQKK208 pKa = 10.01LHH210 pKa = 6.84KK211 pKa = 10.75FPDD214 pKa = 3.55KK215 pKa = 10.52KK216 pKa = 10.76VQPFLTMSNRR226 pKa = 11.84PGIGYY231 pKa = 9.92KK232 pKa = 10.22YY233 pKa = 11.07VLDD236 pKa = 4.17KK237 pKa = 11.47KK238 pKa = 10.16NTLLLTDD245 pKa = 3.81KK246 pKa = 10.84VYY248 pKa = 11.53NNGKK252 pKa = 9.94DD253 pKa = 3.24IPLPRR258 pKa = 11.84YY259 pKa = 7.94YY260 pKa = 10.98LKK262 pKa = 10.89VLEE265 pKa = 4.18NQQYY269 pKa = 10.95CDD271 pKa = 3.26TSTLRR276 pKa = 11.84VNRR279 pKa = 11.84EE280 pKa = 3.65KK281 pKa = 10.93KK282 pKa = 9.81KK283 pKa = 10.84DD284 pKa = 3.91LILEE288 pKa = 4.24EE289 pKa = 4.4EE290 pKa = 4.11NVRR293 pKa = 11.84AYY295 pKa = 9.88KK296 pKa = 9.87LAKK299 pKa = 9.52YY300 pKa = 10.26DD301 pKa = 5.32KK302 pKa = 9.71LLHH305 pKa = 6.06WKK307 pKa = 10.05LEE309 pKa = 4.26KK310 pKa = 10.94GDD312 pKa = 3.73TT313 pKa = 3.72
MM1 pKa = 7.56CVNPIKK7 pKa = 10.37CWQYY11 pKa = 11.4YY12 pKa = 6.65EE13 pKa = 4.34TDD15 pKa = 3.43SLTGEE20 pKa = 4.42EE21 pKa = 4.14KK22 pKa = 9.28THH24 pKa = 7.0LVFNINKK31 pKa = 8.99TSFNKK36 pKa = 10.63DD37 pKa = 2.08KK38 pKa = 10.85DD39 pKa = 3.86YY40 pKa = 11.54SVEE43 pKa = 4.07QKK45 pKa = 11.11LEE47 pKa = 3.84DD48 pKa = 3.84LYY50 pKa = 10.97IPGRR54 pKa = 11.84NQKK57 pKa = 10.18LYY59 pKa = 10.51NRR61 pKa = 11.84RR62 pKa = 11.84IKK64 pKa = 10.5PITLPCGKK72 pKa = 10.38CPDD75 pKa = 4.08CLMKK79 pKa = 10.94YY80 pKa = 9.4SLEE83 pKa = 3.64WAYY86 pKa = 11.05RR87 pKa = 11.84IMNEE91 pKa = 3.3ASYY94 pKa = 10.75YY95 pKa = 10.99AEE97 pKa = 4.13NCFMTLTYY105 pKa = 11.19ANTNGEE111 pKa = 4.37LCLKK115 pKa = 10.36DD116 pKa = 2.97IQNFLKK122 pKa = 10.63RR123 pKa = 11.84LRR125 pKa = 11.84KK126 pKa = 9.78KK127 pKa = 10.5ISPLKK132 pKa = 10.38IRR134 pKa = 11.84FFNCGEE140 pKa = 3.97YY141 pKa = 10.46GSKK144 pKa = 10.27GKK146 pKa = 10.24RR147 pKa = 11.84PHH149 pKa = 5.59FHH151 pKa = 7.07CIIFGWCPSDD161 pKa = 4.85LKK163 pKa = 10.97FLKK166 pKa = 9.02KK167 pKa = 8.76TKK169 pKa = 9.72KK170 pKa = 10.11GQIIYY175 pKa = 9.24TSEE178 pKa = 3.86IVSSLWKK185 pKa = 10.35HH186 pKa = 5.31GFVSIGNVEE195 pKa = 4.19FEE197 pKa = 4.41SAKK200 pKa = 9.4YY201 pKa = 7.37TALYY205 pKa = 7.84MQKK208 pKa = 10.01LHH210 pKa = 6.84KK211 pKa = 10.75FPDD214 pKa = 3.55KK215 pKa = 10.52KK216 pKa = 10.76VQPFLTMSNRR226 pKa = 11.84PGIGYY231 pKa = 9.92KK232 pKa = 10.22YY233 pKa = 11.07VLDD236 pKa = 4.17KK237 pKa = 11.47KK238 pKa = 10.16NTLLLTDD245 pKa = 3.81KK246 pKa = 10.84VYY248 pKa = 11.53NNGKK252 pKa = 9.94DD253 pKa = 3.24IPLPRR258 pKa = 11.84YY259 pKa = 7.94YY260 pKa = 10.98LKK262 pKa = 10.89VLEE265 pKa = 4.18NQQYY269 pKa = 10.95CDD271 pKa = 3.26TSTLRR276 pKa = 11.84VNRR279 pKa = 11.84EE280 pKa = 3.65KK281 pKa = 10.93KK282 pKa = 9.81KK283 pKa = 10.84DD284 pKa = 3.91LILEE288 pKa = 4.24EE289 pKa = 4.4EE290 pKa = 4.11NVRR293 pKa = 11.84AYY295 pKa = 9.88KK296 pKa = 9.87LAKK299 pKa = 9.52YY300 pKa = 10.26DD301 pKa = 5.32KK302 pKa = 9.71LLHH305 pKa = 6.06WKK307 pKa = 10.05LEE309 pKa = 4.26KK310 pKa = 10.94GDD312 pKa = 3.73TT313 pKa = 3.72
Molecular weight: 37.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1609 |
77 |
543 |
201.1 |
22.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.588 ± 1.581 | 1.678 ± 0.581 |
6.091 ± 0.659 | 6.091 ± 1.249 |
4.288 ± 0.474 | 5.594 ± 0.762 |
1.492 ± 0.427 | 5.842 ± 0.649 |
7.396 ± 1.825 | 8.452 ± 0.932 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.175 ± 0.294 | 8.266 ± 0.802 |
3.045 ± 0.951 | 3.418 ± 0.613 |
3.418 ± 0.642 | 7.023 ± 1.02 |
5.158 ± 0.494 | 6.339 ± 0.874 |
1.305 ± 0.163 | 6.339 ± 0.354 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
