
Pseudomonas phage Epa5
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 5.65
Get precalculated fractions of proteins
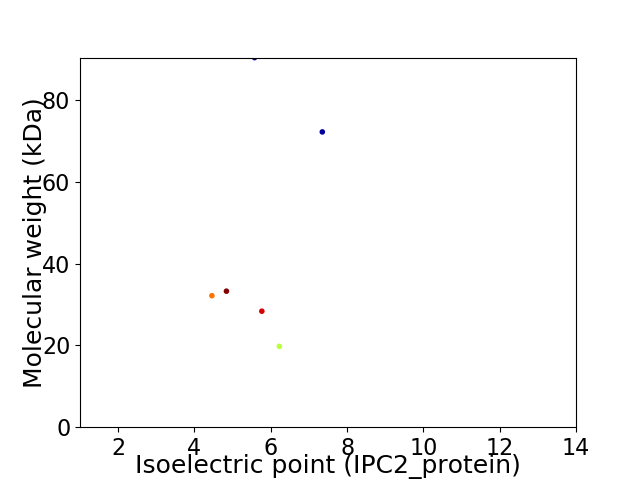
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G9LI61|A0A6G9LI61_9CAUD DNA-directed DNA polymerase OS=Pseudomonas phage Epa5 OX=2719575 GN=Epa5_55 PE=3 SV=1
MM1 pKa = 7.66ASVTLAEE8 pKa = 4.3SAKK11 pKa = 10.49LALDD15 pKa = 3.59EE16 pKa = 4.73LVAGVIEE23 pKa = 4.52NVITVNRR30 pKa = 11.84MFEE33 pKa = 4.34VIPFDD38 pKa = 4.66GISGNALAYY47 pKa = 10.38NRR49 pKa = 11.84EE50 pKa = 4.01NVLGNVQVAGVDD62 pKa = 3.4ATITAKK68 pKa = 10.84APATFTKK75 pKa = 9.51VTSEE79 pKa = 3.93LTTIIGDD86 pKa = 3.74AEE88 pKa = 4.54VNGLIQATRR97 pKa = 11.84SSDD100 pKa = 3.88GNDD103 pKa = 2.98QTAVQVASKK112 pKa = 10.7AKK114 pKa = 9.2SCGRR118 pKa = 11.84KK119 pKa = 8.46YY120 pKa = 11.07QDD122 pKa = 3.21MLINGTGASDD132 pKa = 3.54EE133 pKa = 4.34FEE135 pKa = 5.29GIINLCAEE143 pKa = 4.37GQKK146 pKa = 10.36VATGDD151 pKa = 3.42NGSALSFEE159 pKa = 4.68ILDD162 pKa = 3.73EE163 pKa = 4.74LMDD166 pKa = 4.44LVVDD170 pKa = 4.24KK171 pKa = 10.99DD172 pKa = 3.87GQVDD176 pKa = 4.53YY177 pKa = 9.88LTMHH181 pKa = 6.85ARR183 pKa = 11.84TLRR186 pKa = 11.84SYY188 pKa = 11.04NALLRR193 pKa = 11.84SLGGASIGDD202 pKa = 4.02VVTLPSGAEE211 pKa = 3.83VPAYY215 pKa = 9.93RR216 pKa = 11.84GVPIFRR222 pKa = 11.84NDD224 pKa = 3.92YY225 pKa = 10.8IPTNQTKK232 pKa = 8.19GTGTNTTSIFAGTLDD247 pKa = 4.29DD248 pKa = 4.21GSRR251 pKa = 11.84THH253 pKa = 6.74GLAGLTAEE261 pKa = 4.28EE262 pKa = 3.92AAGIRR267 pKa = 11.84VEE269 pKa = 4.42DD270 pKa = 3.71VGPAEE275 pKa = 5.69DD276 pKa = 3.73KK277 pKa = 11.05DD278 pKa = 4.03NYY280 pKa = 7.42ITRR283 pKa = 11.84VKK285 pKa = 9.32WYY287 pKa = 9.91CGLALFSEE295 pKa = 5.12KK296 pKa = 10.23GLAMADD302 pKa = 4.22GITNN306 pKa = 3.92
MM1 pKa = 7.66ASVTLAEE8 pKa = 4.3SAKK11 pKa = 10.49LALDD15 pKa = 3.59EE16 pKa = 4.73LVAGVIEE23 pKa = 4.52NVITVNRR30 pKa = 11.84MFEE33 pKa = 4.34VIPFDD38 pKa = 4.66GISGNALAYY47 pKa = 10.38NRR49 pKa = 11.84EE50 pKa = 4.01NVLGNVQVAGVDD62 pKa = 3.4ATITAKK68 pKa = 10.84APATFTKK75 pKa = 9.51VTSEE79 pKa = 3.93LTTIIGDD86 pKa = 3.74AEE88 pKa = 4.54VNGLIQATRR97 pKa = 11.84SSDD100 pKa = 3.88GNDD103 pKa = 2.98QTAVQVASKK112 pKa = 10.7AKK114 pKa = 9.2SCGRR118 pKa = 11.84KK119 pKa = 8.46YY120 pKa = 11.07QDD122 pKa = 3.21MLINGTGASDD132 pKa = 3.54EE133 pKa = 4.34FEE135 pKa = 5.29GIINLCAEE143 pKa = 4.37GQKK146 pKa = 10.36VATGDD151 pKa = 3.42NGSALSFEE159 pKa = 4.68ILDD162 pKa = 3.73EE163 pKa = 4.74LMDD166 pKa = 4.44LVVDD170 pKa = 4.24KK171 pKa = 10.99DD172 pKa = 3.87GQVDD176 pKa = 4.53YY177 pKa = 9.88LTMHH181 pKa = 6.85ARR183 pKa = 11.84TLRR186 pKa = 11.84SYY188 pKa = 11.04NALLRR193 pKa = 11.84SLGGASIGDD202 pKa = 4.02VVTLPSGAEE211 pKa = 3.83VPAYY215 pKa = 9.93RR216 pKa = 11.84GVPIFRR222 pKa = 11.84NDD224 pKa = 3.92YY225 pKa = 10.8IPTNQTKK232 pKa = 8.19GTGTNTTSIFAGTLDD247 pKa = 4.29DD248 pKa = 4.21GSRR251 pKa = 11.84THH253 pKa = 6.74GLAGLTAEE261 pKa = 4.28EE262 pKa = 3.92AAGIRR267 pKa = 11.84VEE269 pKa = 4.42DD270 pKa = 3.71VGPAEE275 pKa = 5.69DD276 pKa = 3.73KK277 pKa = 11.05DD278 pKa = 4.03NYY280 pKa = 7.42ITRR283 pKa = 11.84VKK285 pKa = 9.32WYY287 pKa = 9.91CGLALFSEE295 pKa = 5.12KK296 pKa = 10.23GLAMADD302 pKa = 4.22GITNN306 pKa = 3.92
Molecular weight: 32.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G9LI61|A0A6G9LI61_9CAUD DNA-directed DNA polymerase OS=Pseudomonas phage Epa5 OX=2719575 GN=Epa5_55 PE=3 SV=1
MM1 pKa = 6.75MQVPLLTPFSGWKK14 pKa = 10.58APDD17 pKa = 3.72LNALPSWAGHH27 pKa = 5.27KK28 pKa = 10.16RR29 pKa = 11.84IAVDD33 pKa = 4.09CEE35 pKa = 4.06TRR37 pKa = 11.84DD38 pKa = 3.74PFLKK42 pKa = 10.13TLGPGVRR49 pKa = 11.84RR50 pKa = 11.84GGDD53 pKa = 3.18SYY55 pKa = 11.91LVGISFAIEE64 pKa = 4.57DD65 pKa = 4.42GPSGYY70 pKa = 10.68LPIAHH75 pKa = 7.26AGGGNLPKK83 pKa = 10.8EE84 pKa = 4.2MVLKK88 pKa = 10.69YY89 pKa = 10.06MKK91 pKa = 10.52DD92 pKa = 3.06QAAVFDD98 pKa = 4.38GEE100 pKa = 4.36IVGANLQYY108 pKa = 11.38DD109 pKa = 5.02LDD111 pKa = 3.71WLAQYY116 pKa = 10.77GIVFRR121 pKa = 11.84RR122 pKa = 11.84AKK124 pKa = 10.2YY125 pKa = 10.03FRR127 pKa = 11.84DD128 pKa = 3.66VQVAEE133 pKa = 4.12PLIDD137 pKa = 4.0EE138 pKa = 4.78LQLSYY143 pKa = 11.57SLDD146 pKa = 4.1NIAKK150 pKa = 9.08RR151 pKa = 11.84HH152 pKa = 5.54GIAGKK157 pKa = 10.35DD158 pKa = 3.35EE159 pKa = 4.23TLLMQAARR167 pKa = 11.84AYY169 pKa = 10.13GIHH172 pKa = 6.85PKK174 pKa = 10.01KK175 pKa = 10.7DD176 pKa = 3.17LWRR179 pKa = 11.84LPAQFVGPYY188 pKa = 9.79AEE190 pKa = 4.21EE191 pKa = 4.33DD192 pKa = 3.3ARR194 pKa = 11.84LPLQLLRR201 pKa = 11.84RR202 pKa = 11.84QEE204 pKa = 4.38RR205 pKa = 11.84IIDD208 pKa = 3.87DD209 pKa = 3.64QEE211 pKa = 3.4IWDD214 pKa = 4.13IYY216 pKa = 10.11NLEE219 pKa = 4.4SKK221 pKa = 10.35VLPVLVKK228 pKa = 8.76MRR230 pKa = 11.84RR231 pKa = 11.84RR232 pKa = 11.84GVRR235 pKa = 11.84VHH237 pKa = 6.83EE238 pKa = 4.58GRR240 pKa = 11.84LTEE243 pKa = 3.82IEE245 pKa = 4.13RR246 pKa = 11.84WSLQQEE252 pKa = 4.49TEE254 pKa = 3.6ALAQVRR260 pKa = 11.84HH261 pKa = 4.8LTGVNIAVGDD271 pKa = 3.67VWKK274 pKa = 11.21NEE276 pKa = 3.73ALAPALTHH284 pKa = 7.28IGLKK288 pKa = 10.18LNKK291 pKa = 8.46TAEE294 pKa = 4.45GKK296 pKa = 10.67VSIDD300 pKa = 3.26KK301 pKa = 10.75EE302 pKa = 4.02FLAGVDD308 pKa = 3.36HH309 pKa = 7.44DD310 pKa = 3.94VARR313 pKa = 11.84ALEE316 pKa = 4.16RR317 pKa = 11.84ARR319 pKa = 11.84KK320 pKa = 6.8TNKK323 pKa = 9.74LRR325 pKa = 11.84TTFAASVRR333 pKa = 11.84EE334 pKa = 3.85HH335 pKa = 5.78MVNGRR340 pKa = 11.84IHH342 pKa = 6.47ATFNQLRR349 pKa = 11.84MEE351 pKa = 4.89KK352 pKa = 10.76DD353 pKa = 3.09NGDD356 pKa = 3.82LGGAAYY362 pKa = 10.06GRR364 pKa = 11.84LSCVQPNLQQQPARR378 pKa = 11.84DD379 pKa = 4.23DD380 pKa = 3.69FAKK383 pKa = 8.7MWRR386 pKa = 11.84GIYY389 pKa = 10.17LPEE392 pKa = 4.96EE393 pKa = 4.4GQLWAALDD401 pKa = 3.92YY402 pKa = 10.73SQQEE406 pKa = 3.97PRR408 pKa = 11.84MTVHH412 pKa = 6.99FAEE415 pKa = 4.76LCGLPGSKK423 pKa = 9.93EE424 pKa = 3.91AGDD427 pKa = 4.57KK428 pKa = 10.99YY429 pKa = 10.26RR430 pKa = 11.84TDD432 pKa = 3.86PNADD436 pKa = 3.33NHH438 pKa = 6.83QMMADD443 pKa = 3.3MAKK446 pKa = 9.59IQRR449 pKa = 11.84KK450 pKa = 6.72AAKK453 pKa = 9.77EE454 pKa = 3.77IFLGLCYY461 pKa = 10.67GMGGAKK467 pKa = 10.15LCRR470 pKa = 11.84KK471 pKa = 9.83LGLPTAWAVQHH482 pKa = 6.29RR483 pKa = 11.84NIRR486 pKa = 11.84GLVALDD492 pKa = 3.46TEE494 pKa = 5.03RR495 pKa = 11.84GQEE498 pKa = 4.18LLRR501 pKa = 11.84QGGKK505 pKa = 9.73KK506 pKa = 10.33LEE508 pKa = 4.09IAGPEE513 pKa = 4.22GQALLDD519 pKa = 3.71TFDD522 pKa = 3.87AKK524 pKa = 11.29VPFVRR529 pKa = 11.84QLARR533 pKa = 11.84RR534 pKa = 11.84CSKK537 pKa = 9.93VAAEE541 pKa = 4.38RR542 pKa = 11.84GYY544 pKa = 11.58LMTLAGRR551 pKa = 11.84RR552 pKa = 11.84CRR554 pKa = 11.84FPKK557 pKa = 10.09TPEE560 pKa = 3.69GKK562 pKa = 10.36YY563 pKa = 10.37DD564 pKa = 3.5WTHH567 pKa = 6.21KK568 pKa = 10.48GLNRR572 pKa = 11.84LIQGSSGDD580 pKa = 3.62QTKK583 pKa = 8.35TAMVMLDD590 pKa = 3.46EE591 pKa = 5.28AGHH594 pKa = 5.34YY595 pKa = 10.02LQLQVHH601 pKa = 6.99DD602 pKa = 4.57EE603 pKa = 4.24MDD605 pKa = 3.74GSVQSPEE612 pKa = 3.82EE613 pKa = 4.36AEE615 pKa = 4.04AMAEE619 pKa = 3.99IMRR622 pKa = 11.84TCTPLTVPSKK632 pKa = 11.28VDD634 pKa = 3.44VEE636 pKa = 5.45LGASWGEE643 pKa = 4.25SMSS646 pKa = 3.5
MM1 pKa = 6.75MQVPLLTPFSGWKK14 pKa = 10.58APDD17 pKa = 3.72LNALPSWAGHH27 pKa = 5.27KK28 pKa = 10.16RR29 pKa = 11.84IAVDD33 pKa = 4.09CEE35 pKa = 4.06TRR37 pKa = 11.84DD38 pKa = 3.74PFLKK42 pKa = 10.13TLGPGVRR49 pKa = 11.84RR50 pKa = 11.84GGDD53 pKa = 3.18SYY55 pKa = 11.91LVGISFAIEE64 pKa = 4.57DD65 pKa = 4.42GPSGYY70 pKa = 10.68LPIAHH75 pKa = 7.26AGGGNLPKK83 pKa = 10.8EE84 pKa = 4.2MVLKK88 pKa = 10.69YY89 pKa = 10.06MKK91 pKa = 10.52DD92 pKa = 3.06QAAVFDD98 pKa = 4.38GEE100 pKa = 4.36IVGANLQYY108 pKa = 11.38DD109 pKa = 5.02LDD111 pKa = 3.71WLAQYY116 pKa = 10.77GIVFRR121 pKa = 11.84RR122 pKa = 11.84AKK124 pKa = 10.2YY125 pKa = 10.03FRR127 pKa = 11.84DD128 pKa = 3.66VQVAEE133 pKa = 4.12PLIDD137 pKa = 4.0EE138 pKa = 4.78LQLSYY143 pKa = 11.57SLDD146 pKa = 4.1NIAKK150 pKa = 9.08RR151 pKa = 11.84HH152 pKa = 5.54GIAGKK157 pKa = 10.35DD158 pKa = 3.35EE159 pKa = 4.23TLLMQAARR167 pKa = 11.84AYY169 pKa = 10.13GIHH172 pKa = 6.85PKK174 pKa = 10.01KK175 pKa = 10.7DD176 pKa = 3.17LWRR179 pKa = 11.84LPAQFVGPYY188 pKa = 9.79AEE190 pKa = 4.21EE191 pKa = 4.33DD192 pKa = 3.3ARR194 pKa = 11.84LPLQLLRR201 pKa = 11.84RR202 pKa = 11.84QEE204 pKa = 4.38RR205 pKa = 11.84IIDD208 pKa = 3.87DD209 pKa = 3.64QEE211 pKa = 3.4IWDD214 pKa = 4.13IYY216 pKa = 10.11NLEE219 pKa = 4.4SKK221 pKa = 10.35VLPVLVKK228 pKa = 8.76MRR230 pKa = 11.84RR231 pKa = 11.84RR232 pKa = 11.84GVRR235 pKa = 11.84VHH237 pKa = 6.83EE238 pKa = 4.58GRR240 pKa = 11.84LTEE243 pKa = 3.82IEE245 pKa = 4.13RR246 pKa = 11.84WSLQQEE252 pKa = 4.49TEE254 pKa = 3.6ALAQVRR260 pKa = 11.84HH261 pKa = 4.8LTGVNIAVGDD271 pKa = 3.67VWKK274 pKa = 11.21NEE276 pKa = 3.73ALAPALTHH284 pKa = 7.28IGLKK288 pKa = 10.18LNKK291 pKa = 8.46TAEE294 pKa = 4.45GKK296 pKa = 10.67VSIDD300 pKa = 3.26KK301 pKa = 10.75EE302 pKa = 4.02FLAGVDD308 pKa = 3.36HH309 pKa = 7.44DD310 pKa = 3.94VARR313 pKa = 11.84ALEE316 pKa = 4.16RR317 pKa = 11.84ARR319 pKa = 11.84KK320 pKa = 6.8TNKK323 pKa = 9.74LRR325 pKa = 11.84TTFAASVRR333 pKa = 11.84EE334 pKa = 3.85HH335 pKa = 5.78MVNGRR340 pKa = 11.84IHH342 pKa = 6.47ATFNQLRR349 pKa = 11.84MEE351 pKa = 4.89KK352 pKa = 10.76DD353 pKa = 3.09NGDD356 pKa = 3.82LGGAAYY362 pKa = 10.06GRR364 pKa = 11.84LSCVQPNLQQQPARR378 pKa = 11.84DD379 pKa = 4.23DD380 pKa = 3.69FAKK383 pKa = 8.7MWRR386 pKa = 11.84GIYY389 pKa = 10.17LPEE392 pKa = 4.96EE393 pKa = 4.4GQLWAALDD401 pKa = 3.92YY402 pKa = 10.73SQQEE406 pKa = 3.97PRR408 pKa = 11.84MTVHH412 pKa = 6.99FAEE415 pKa = 4.76LCGLPGSKK423 pKa = 9.93EE424 pKa = 3.91AGDD427 pKa = 4.57KK428 pKa = 10.99YY429 pKa = 10.26RR430 pKa = 11.84TDD432 pKa = 3.86PNADD436 pKa = 3.33NHH438 pKa = 6.83QMMADD443 pKa = 3.3MAKK446 pKa = 9.59IQRR449 pKa = 11.84KK450 pKa = 6.72AAKK453 pKa = 9.77EE454 pKa = 3.77IFLGLCYY461 pKa = 10.67GMGGAKK467 pKa = 10.15LCRR470 pKa = 11.84KK471 pKa = 9.83LGLPTAWAVQHH482 pKa = 6.29RR483 pKa = 11.84NIRR486 pKa = 11.84GLVALDD492 pKa = 3.46TEE494 pKa = 5.03RR495 pKa = 11.84GQEE498 pKa = 4.18LLRR501 pKa = 11.84QGGKK505 pKa = 9.73KK506 pKa = 10.33LEE508 pKa = 4.09IAGPEE513 pKa = 4.22GQALLDD519 pKa = 3.71TFDD522 pKa = 3.87AKK524 pKa = 11.29VPFVRR529 pKa = 11.84QLARR533 pKa = 11.84RR534 pKa = 11.84CSKK537 pKa = 9.93VAAEE541 pKa = 4.38RR542 pKa = 11.84GYY544 pKa = 11.58LMTLAGRR551 pKa = 11.84RR552 pKa = 11.84CRR554 pKa = 11.84FPKK557 pKa = 10.09TPEE560 pKa = 3.69GKK562 pKa = 10.36YY563 pKa = 10.37DD564 pKa = 3.5WTHH567 pKa = 6.21KK568 pKa = 10.48GLNRR572 pKa = 11.84LIQGSSGDD580 pKa = 3.62QTKK583 pKa = 8.35TAMVMLDD590 pKa = 3.46EE591 pKa = 5.28AGHH594 pKa = 5.34YY595 pKa = 10.02LQLQVHH601 pKa = 6.99DD602 pKa = 4.57EE603 pKa = 4.24MDD605 pKa = 3.74GSVQSPEE612 pKa = 3.82EE613 pKa = 4.36AEE615 pKa = 4.04AMAEE619 pKa = 3.99IMRR622 pKa = 11.84TCTPLTVPSKK632 pKa = 11.28VDD634 pKa = 3.44VEE636 pKa = 5.45LGASWGEE643 pKa = 4.25SMSS646 pKa = 3.5
Molecular weight: 72.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2495 |
180 |
790 |
415.8 |
46.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.499 ± 0.762 | 0.882 ± 0.179 |
6.573 ± 0.411 | 7.174 ± 0.497 |
3.768 ± 0.485 | 8.617 ± 0.498 |
1.924 ± 0.473 | 4.489 ± 0.47 |
6.052 ± 0.749 | 8.858 ± 0.698 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.645 ± 0.342 | 3.567 ± 0.434 |
4.81 ± 0.457 | 4.329 ± 0.469 |
5.972 ± 0.546 | 4.97 ± 0.651 |
5.13 ± 1.203 | 6.653 ± 0.372 |
1.242 ± 0.38 | 2.846 ± 0.197 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
