
Rousettus aegyptiacus papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Psipapillomavirus; Psipapillomavirus 1
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
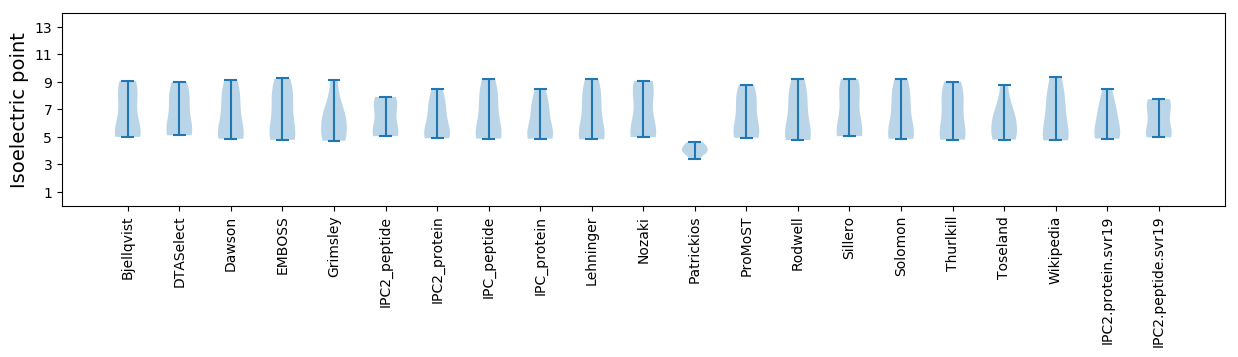
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q0QII1|Q0QII1_9PAPI Protein E6 OS=Rousettus aegyptiacus papillomavirus 1 OX=369584 GN=E6 PE=3 SV=1
MM1 pKa = 8.02RR2 pKa = 11.84GRR4 pKa = 11.84DD5 pKa = 3.61PSLEE9 pKa = 3.76LAAAAEE15 pKa = 4.34EE16 pKa = 4.39VQAVDD21 pKa = 3.54LHH23 pKa = 7.8CDD25 pKa = 3.17EE26 pKa = 5.91SLDD29 pKa = 4.66QEE31 pKa = 5.29AEE33 pKa = 3.79QQTFYY38 pKa = 11.23NIQLPCTSCEE48 pKa = 3.3RR49 pKa = 11.84HH50 pKa = 4.9MRR52 pKa = 11.84LVISCSSSGLRR63 pKa = 11.84TLAVLLRR70 pKa = 11.84QRR72 pKa = 11.84EE73 pKa = 4.15VQLICAQCTRR83 pKa = 11.84EE84 pKa = 3.84FHH86 pKa = 6.54
MM1 pKa = 8.02RR2 pKa = 11.84GRR4 pKa = 11.84DD5 pKa = 3.61PSLEE9 pKa = 3.76LAAAAEE15 pKa = 4.34EE16 pKa = 4.39VQAVDD21 pKa = 3.54LHH23 pKa = 7.8CDD25 pKa = 3.17EE26 pKa = 5.91SLDD29 pKa = 4.66QEE31 pKa = 5.29AEE33 pKa = 3.79QQTFYY38 pKa = 11.23NIQLPCTSCEE48 pKa = 3.3RR49 pKa = 11.84HH50 pKa = 4.9MRR52 pKa = 11.84LVISCSSSGLRR63 pKa = 11.84TLAVLLRR70 pKa = 11.84QRR72 pKa = 11.84EE73 pKa = 4.15VQLICAQCTRR83 pKa = 11.84EE84 pKa = 3.84FHH86 pKa = 6.54
Molecular weight: 9.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q0QIH9|Q0QIH9_9PAPI Replication protein E1 OS=Rousettus aegyptiacus papillomavirus 1 OX=369584 GN=E1 PE=3 SV=1
MM1 pKa = 6.94EE2 pKa = 4.74TLRR5 pKa = 11.84DD6 pKa = 3.87RR7 pKa = 11.84LDD9 pKa = 3.39VLQDD13 pKa = 3.47QILGHH18 pKa = 5.81YY19 pKa = 7.6EE20 pKa = 3.9RR21 pKa = 11.84QSTCLQDD28 pKa = 4.14HH29 pKa = 4.87VQYY32 pKa = 9.77WGLVRR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 3.97SSLLFYY45 pKa = 11.15ARR47 pKa = 11.84GKK49 pKa = 9.63GVKK52 pKa = 8.53TLGYY56 pKa = 10.33LPVPVQQVSQDD67 pKa = 3.28RR68 pKa = 11.84ARR70 pKa = 11.84QAIQLHH76 pKa = 6.69LATQSLAKK84 pKa = 9.93SKK86 pKa = 11.22YY87 pKa = 9.53KK88 pKa = 10.54DD89 pKa = 3.96EE90 pKa = 4.52PWTLQDD96 pKa = 3.46VSRR99 pKa = 11.84EE100 pKa = 4.02NYY102 pKa = 8.98MSEE105 pKa = 4.04PKK107 pKa = 9.13HH108 pKa = 4.91TFKK111 pKa = 11.03KK112 pKa = 10.49GGTQVEE118 pKa = 4.62VIYY121 pKa = 11.09DD122 pKa = 3.64GDD124 pKa = 3.83EE125 pKa = 4.55DD126 pKa = 4.79NVMLYY131 pKa = 9.54TSWEE135 pKa = 4.09HH136 pKa = 8.36IYY138 pKa = 10.51IQDD141 pKa = 5.6DD142 pKa = 3.55DD143 pKa = 4.97GSWYY147 pKa = 9.47KK148 pKa = 10.62RR149 pKa = 11.84KK150 pKa = 10.73GEE152 pKa = 4.04VCVEE156 pKa = 3.66GLYY159 pKa = 8.21YY160 pKa = 10.22TWLGQKK166 pKa = 7.79TFYY169 pKa = 11.51VNFEE173 pKa = 4.28EE174 pKa = 4.9EE175 pKa = 4.02LGKK178 pKa = 10.96YY179 pKa = 7.11STRR182 pKa = 11.84NVYY185 pKa = 8.39VVRR188 pKa = 11.84TADD191 pKa = 3.55GEE193 pKa = 4.39LMSFNPVTSSTPQEE207 pKa = 4.38EE208 pKa = 4.85GQQPGGGAPGHH219 pKa = 6.76PGPPPRR225 pKa = 11.84KK226 pKa = 8.45RR227 pKa = 11.84RR228 pKa = 11.84RR229 pKa = 11.84LFGEE233 pKa = 4.51KK234 pKa = 9.38PAEE237 pKa = 4.39GPGGRR242 pKa = 11.84RR243 pKa = 11.84VPVPRR248 pKa = 11.84PGGPPPGRR256 pKa = 11.84RR257 pKa = 11.84GGEE260 pKa = 4.0TPSRR264 pKa = 11.84GTEE267 pKa = 3.82RR268 pKa = 11.84PAEE271 pKa = 4.18GSGGCIPPALPGAAGKK287 pKa = 8.19TEE289 pKa = 3.85EE290 pKa = 4.37RR291 pKa = 11.84GGAGFRR297 pKa = 11.84PQPPTAVVAPVIIIAGPPNALKK319 pKa = 10.08CQRR322 pKa = 11.84YY323 pKa = 8.82RR324 pKa = 11.84LQNQHH329 pKa = 5.57YY330 pKa = 8.98RR331 pKa = 11.84HH332 pKa = 6.33FAQVTTTYY340 pKa = 9.54HH341 pKa = 6.15WLDD344 pKa = 3.43AGHH347 pKa = 5.15TQRR350 pKa = 11.84RR351 pKa = 11.84SAAQIVLRR359 pKa = 11.84FVDD362 pKa = 3.86TAQRR366 pKa = 11.84EE367 pKa = 4.55TFLNTGALSDD377 pKa = 3.63RR378 pKa = 11.84VHH380 pKa = 6.14YY381 pKa = 10.8VLGTMPFLSS390 pKa = 3.86
MM1 pKa = 6.94EE2 pKa = 4.74TLRR5 pKa = 11.84DD6 pKa = 3.87RR7 pKa = 11.84LDD9 pKa = 3.39VLQDD13 pKa = 3.47QILGHH18 pKa = 5.81YY19 pKa = 7.6EE20 pKa = 3.9RR21 pKa = 11.84QSTCLQDD28 pKa = 4.14HH29 pKa = 4.87VQYY32 pKa = 9.77WGLVRR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 3.97SSLLFYY45 pKa = 11.15ARR47 pKa = 11.84GKK49 pKa = 9.63GVKK52 pKa = 8.53TLGYY56 pKa = 10.33LPVPVQQVSQDD67 pKa = 3.28RR68 pKa = 11.84ARR70 pKa = 11.84QAIQLHH76 pKa = 6.69LATQSLAKK84 pKa = 9.93SKK86 pKa = 11.22YY87 pKa = 9.53KK88 pKa = 10.54DD89 pKa = 3.96EE90 pKa = 4.52PWTLQDD96 pKa = 3.46VSRR99 pKa = 11.84EE100 pKa = 4.02NYY102 pKa = 8.98MSEE105 pKa = 4.04PKK107 pKa = 9.13HH108 pKa = 4.91TFKK111 pKa = 11.03KK112 pKa = 10.49GGTQVEE118 pKa = 4.62VIYY121 pKa = 11.09DD122 pKa = 3.64GDD124 pKa = 3.83EE125 pKa = 4.55DD126 pKa = 4.79NVMLYY131 pKa = 9.54TSWEE135 pKa = 4.09HH136 pKa = 8.36IYY138 pKa = 10.51IQDD141 pKa = 5.6DD142 pKa = 3.55DD143 pKa = 4.97GSWYY147 pKa = 9.47KK148 pKa = 10.62RR149 pKa = 11.84KK150 pKa = 10.73GEE152 pKa = 4.04VCVEE156 pKa = 3.66GLYY159 pKa = 8.21YY160 pKa = 10.22TWLGQKK166 pKa = 7.79TFYY169 pKa = 11.51VNFEE173 pKa = 4.28EE174 pKa = 4.9EE175 pKa = 4.02LGKK178 pKa = 10.96YY179 pKa = 7.11STRR182 pKa = 11.84NVYY185 pKa = 8.39VVRR188 pKa = 11.84TADD191 pKa = 3.55GEE193 pKa = 4.39LMSFNPVTSSTPQEE207 pKa = 4.38EE208 pKa = 4.85GQQPGGGAPGHH219 pKa = 6.76PGPPPRR225 pKa = 11.84KK226 pKa = 8.45RR227 pKa = 11.84RR228 pKa = 11.84RR229 pKa = 11.84LFGEE233 pKa = 4.51KK234 pKa = 9.38PAEE237 pKa = 4.39GPGGRR242 pKa = 11.84RR243 pKa = 11.84VPVPRR248 pKa = 11.84PGGPPPGRR256 pKa = 11.84RR257 pKa = 11.84GGEE260 pKa = 4.0TPSRR264 pKa = 11.84GTEE267 pKa = 3.82RR268 pKa = 11.84PAEE271 pKa = 4.18GSGGCIPPALPGAAGKK287 pKa = 8.19TEE289 pKa = 3.85EE290 pKa = 4.37RR291 pKa = 11.84GGAGFRR297 pKa = 11.84PQPPTAVVAPVIIIAGPPNALKK319 pKa = 10.08CQRR322 pKa = 11.84YY323 pKa = 8.82RR324 pKa = 11.84LQNQHH329 pKa = 5.57YY330 pKa = 8.98RR331 pKa = 11.84HH332 pKa = 6.33FAQVTTTYY340 pKa = 9.54HH341 pKa = 6.15WLDD344 pKa = 3.43AGHH347 pKa = 5.15TQRR350 pKa = 11.84RR351 pKa = 11.84SAAQIVLRR359 pKa = 11.84FVDD362 pKa = 3.86TAQRR366 pKa = 11.84EE367 pKa = 4.55TFLNTGALSDD377 pKa = 3.63RR378 pKa = 11.84VHH380 pKa = 6.14YY381 pKa = 10.8VLGTMPFLSS390 pKa = 3.86
Molecular weight: 43.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2507 |
86 |
665 |
358.1 |
39.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.781 ± 0.499 | 2.074 ± 0.472 |
6.143 ± 0.432 | 6.502 ± 0.44 |
3.151 ± 0.332 | 8.456 ± 0.571 |
2.513 ± 0.26 | 4.428 ± 0.662 |
3.949 ± 0.54 | 8.217 ± 0.551 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.034 ± 0.363 | 3.032 ± 0.632 |
7.06 ± 0.94 | 4.826 ± 0.658 |
6.781 ± 0.641 | 6.661 ± 0.734 |
5.943 ± 0.585 | 7.06 ± 0.468 |
1.197 ± 0.205 | 3.191 ± 0.356 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
