
Lake Sarah-associated circular molecule 9
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.68
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126GA21|A0A126GA21_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular molecule 9 OX=1685734 PE=3 SV=1
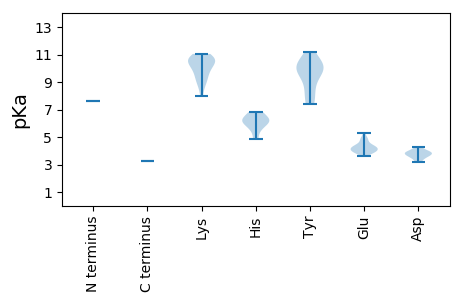
MM1 pKa = 7.62SKK3 pKa = 10.85GNRR6 pKa = 11.84AWCYY10 pKa = 8.56TLNNYY15 pKa = 7.52TEE17 pKa = 4.32EE18 pKa = 4.27EE19 pKa = 4.09RR20 pKa = 11.84DD21 pKa = 3.48SLRR24 pKa = 11.84SLKK27 pKa = 10.3CAYY30 pKa = 9.82QVFGYY35 pKa = 10.34EE36 pKa = 3.98RR37 pKa = 11.84GAADD41 pKa = 3.75TPHH44 pKa = 6.32LQGYY48 pKa = 7.38VQFAHH53 pKa = 6.15QKK55 pKa = 8.0TLSAVKK61 pKa = 10.22KK62 pKa = 9.18LLPRR66 pKa = 11.84AHH68 pKa = 6.81LEE70 pKa = 3.97EE71 pKa = 4.21RR72 pKa = 11.84RR73 pKa = 11.84GTIDD77 pKa = 4.26QAVEE81 pKa = 3.96YY82 pKa = 9.63CKK84 pKa = 10.52KK85 pKa = 10.79DD86 pKa = 3.19GDD88 pKa = 3.81FEE90 pKa = 4.99EE91 pKa = 4.82YY92 pKa = 10.34GKK94 pKa = 11.05KK95 pKa = 10.16PMSQKK100 pKa = 10.71EE101 pKa = 3.93KK102 pKa = 10.88GKK104 pKa = 9.76EE105 pKa = 3.71EE106 pKa = 3.87KK107 pKa = 10.55NRR109 pKa = 11.84WKK111 pKa = 10.7RR112 pKa = 11.84ILEE115 pKa = 4.21KK116 pKa = 10.47ADD118 pKa = 3.58EE119 pKa = 4.81GDD121 pKa = 3.93EE122 pKa = 3.88EE123 pKa = 4.33WLRR126 pKa = 11.84EE127 pKa = 4.02NEE129 pKa = 3.8PNVAFKK135 pKa = 10.97HH136 pKa = 4.83MATFRR141 pKa = 11.84SHH143 pKa = 6.76KK144 pKa = 9.34KK145 pKa = 9.42PRR147 pKa = 11.84VGTLQYY153 pKa = 11.2EE154 pKa = 4.54EE155 pKa = 5.3TPHH158 pKa = 5.8EE159 pKa = 4.37WWVGPTGTGKK169 pKa = 9.97SRR171 pKa = 11.84KK172 pKa = 8.83AHH174 pKa = 5.54EE175 pKa = 4.93EE176 pKa = 4.26YY177 pKa = 9.97PNHH180 pKa = 6.13YY181 pKa = 10.49AKK183 pKa = 10.6EE184 pKa = 4.11KK185 pKa = 9.03NKK187 pKa = 8.6WWCGYY192 pKa = 8.35TGQEE196 pKa = 3.87TVIIEE201 pKa = 4.12EE202 pKa = 4.5ADD204 pKa = 3.7PKK206 pKa = 10.71TMEE209 pKa = 4.43HH210 pKa = 6.23LAARR214 pKa = 11.84LKK216 pKa = 10.41VWADD220 pKa = 3.92RR221 pKa = 11.84YY222 pKa = 9.85PFPGEE227 pKa = 3.63IKK229 pKa = 10.02GGRR232 pKa = 11.84IEE234 pKa = 4.69GIRR237 pKa = 11.84PLRR240 pKa = 11.84VIVISNYY247 pKa = 9.56TIEE250 pKa = 4.12EE251 pKa = 4.25CFANQNDD258 pKa = 3.94VEE260 pKa = 4.16PLRR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84FKK267 pKa = 9.93EE268 pKa = 3.96VKK270 pKa = 9.55FGEE273 pKa = 4.33RR274 pKa = 11.84TMPSPWHH281 pKa = 6.22PSYY284 pKa = 10.29TLL286 pKa = 3.27
MM1 pKa = 7.62SKK3 pKa = 10.85GNRR6 pKa = 11.84AWCYY10 pKa = 8.56TLNNYY15 pKa = 7.52TEE17 pKa = 4.32EE18 pKa = 4.27EE19 pKa = 4.09RR20 pKa = 11.84DD21 pKa = 3.48SLRR24 pKa = 11.84SLKK27 pKa = 10.3CAYY30 pKa = 9.82QVFGYY35 pKa = 10.34EE36 pKa = 3.98RR37 pKa = 11.84GAADD41 pKa = 3.75TPHH44 pKa = 6.32LQGYY48 pKa = 7.38VQFAHH53 pKa = 6.15QKK55 pKa = 8.0TLSAVKK61 pKa = 10.22KK62 pKa = 9.18LLPRR66 pKa = 11.84AHH68 pKa = 6.81LEE70 pKa = 3.97EE71 pKa = 4.21RR72 pKa = 11.84RR73 pKa = 11.84GTIDD77 pKa = 4.26QAVEE81 pKa = 3.96YY82 pKa = 9.63CKK84 pKa = 10.52KK85 pKa = 10.79DD86 pKa = 3.19GDD88 pKa = 3.81FEE90 pKa = 4.99EE91 pKa = 4.82YY92 pKa = 10.34GKK94 pKa = 11.05KK95 pKa = 10.16PMSQKK100 pKa = 10.71EE101 pKa = 3.93KK102 pKa = 10.88GKK104 pKa = 9.76EE105 pKa = 3.71EE106 pKa = 3.87KK107 pKa = 10.55NRR109 pKa = 11.84WKK111 pKa = 10.7RR112 pKa = 11.84ILEE115 pKa = 4.21KK116 pKa = 10.47ADD118 pKa = 3.58EE119 pKa = 4.81GDD121 pKa = 3.93EE122 pKa = 3.88EE123 pKa = 4.33WLRR126 pKa = 11.84EE127 pKa = 4.02NEE129 pKa = 3.8PNVAFKK135 pKa = 10.97HH136 pKa = 4.83MATFRR141 pKa = 11.84SHH143 pKa = 6.76KK144 pKa = 9.34KK145 pKa = 9.42PRR147 pKa = 11.84VGTLQYY153 pKa = 11.2EE154 pKa = 4.54EE155 pKa = 5.3TPHH158 pKa = 5.8EE159 pKa = 4.37WWVGPTGTGKK169 pKa = 9.97SRR171 pKa = 11.84KK172 pKa = 8.83AHH174 pKa = 5.54EE175 pKa = 4.93EE176 pKa = 4.26YY177 pKa = 9.97PNHH180 pKa = 6.13YY181 pKa = 10.49AKK183 pKa = 10.6EE184 pKa = 4.11KK185 pKa = 9.03NKK187 pKa = 8.6WWCGYY192 pKa = 8.35TGQEE196 pKa = 3.87TVIIEE201 pKa = 4.12EE202 pKa = 4.5ADD204 pKa = 3.7PKK206 pKa = 10.71TMEE209 pKa = 4.43HH210 pKa = 6.23LAARR214 pKa = 11.84LKK216 pKa = 10.41VWADD220 pKa = 3.92RR221 pKa = 11.84YY222 pKa = 9.85PFPGEE227 pKa = 3.63IKK229 pKa = 10.02GGRR232 pKa = 11.84IEE234 pKa = 4.69GIRR237 pKa = 11.84PLRR240 pKa = 11.84VIVISNYY247 pKa = 9.56TIEE250 pKa = 4.12EE251 pKa = 4.25CFANQNDD258 pKa = 3.94VEE260 pKa = 4.16PLRR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84FKK267 pKa = 9.93EE268 pKa = 3.96VKK270 pKa = 9.55FGEE273 pKa = 4.33RR274 pKa = 11.84TMPSPWHH281 pKa = 6.22PSYY284 pKa = 10.29TLL286 pKa = 3.27
Molecular weight: 33.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GA21|A0A126GA21_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular molecule 9 OX=1685734 PE=3 SV=1
MM1 pKa = 7.62SKK3 pKa = 10.85GNRR6 pKa = 11.84AWCYY10 pKa = 8.56TLNNYY15 pKa = 7.52TEE17 pKa = 4.32EE18 pKa = 4.27EE19 pKa = 4.09RR20 pKa = 11.84DD21 pKa = 3.48SLRR24 pKa = 11.84SLKK27 pKa = 10.3CAYY30 pKa = 9.82QVFGYY35 pKa = 10.34EE36 pKa = 3.98RR37 pKa = 11.84GAADD41 pKa = 3.75TPHH44 pKa = 6.32LQGYY48 pKa = 7.38VQFAHH53 pKa = 6.15QKK55 pKa = 8.0TLSAVKK61 pKa = 10.22KK62 pKa = 9.18LLPRR66 pKa = 11.84AHH68 pKa = 6.81LEE70 pKa = 3.97EE71 pKa = 4.21RR72 pKa = 11.84RR73 pKa = 11.84GTIDD77 pKa = 4.26QAVEE81 pKa = 3.96YY82 pKa = 9.63CKK84 pKa = 10.52KK85 pKa = 10.79DD86 pKa = 3.19GDD88 pKa = 3.81FEE90 pKa = 4.99EE91 pKa = 4.82YY92 pKa = 10.34GKK94 pKa = 11.05KK95 pKa = 10.16PMSQKK100 pKa = 10.71EE101 pKa = 3.93KK102 pKa = 10.88GKK104 pKa = 9.76EE105 pKa = 3.71EE106 pKa = 3.87KK107 pKa = 10.55NRR109 pKa = 11.84WKK111 pKa = 10.7RR112 pKa = 11.84ILEE115 pKa = 4.21KK116 pKa = 10.47ADD118 pKa = 3.58EE119 pKa = 4.81GDD121 pKa = 3.93EE122 pKa = 3.88EE123 pKa = 4.33WLRR126 pKa = 11.84EE127 pKa = 4.02NEE129 pKa = 3.8PNVAFKK135 pKa = 10.97HH136 pKa = 4.83MATFRR141 pKa = 11.84SHH143 pKa = 6.76KK144 pKa = 9.34KK145 pKa = 9.42PRR147 pKa = 11.84VGTLQYY153 pKa = 11.2EE154 pKa = 4.54EE155 pKa = 5.3TPHH158 pKa = 5.8EE159 pKa = 4.37WWVGPTGTGKK169 pKa = 9.97SRR171 pKa = 11.84KK172 pKa = 8.83AHH174 pKa = 5.54EE175 pKa = 4.93EE176 pKa = 4.26YY177 pKa = 9.97PNHH180 pKa = 6.13YY181 pKa = 10.49AKK183 pKa = 10.6EE184 pKa = 4.11KK185 pKa = 9.03NKK187 pKa = 8.6WWCGYY192 pKa = 8.35TGQEE196 pKa = 3.87TVIIEE201 pKa = 4.12EE202 pKa = 4.5ADD204 pKa = 3.7PKK206 pKa = 10.71TMEE209 pKa = 4.43HH210 pKa = 6.23LAARR214 pKa = 11.84LKK216 pKa = 10.41VWADD220 pKa = 3.92RR221 pKa = 11.84YY222 pKa = 9.85PFPGEE227 pKa = 3.63IKK229 pKa = 10.02GGRR232 pKa = 11.84IEE234 pKa = 4.69GIRR237 pKa = 11.84PLRR240 pKa = 11.84VIVISNYY247 pKa = 9.56TIEE250 pKa = 4.12EE251 pKa = 4.25CFANQNDD258 pKa = 3.94VEE260 pKa = 4.16PLRR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84FKK267 pKa = 9.93EE268 pKa = 3.96VKK270 pKa = 9.55FGEE273 pKa = 4.33RR274 pKa = 11.84TMPSPWHH281 pKa = 6.22PSYY284 pKa = 10.29TLL286 pKa = 3.27
MM1 pKa = 7.62SKK3 pKa = 10.85GNRR6 pKa = 11.84AWCYY10 pKa = 8.56TLNNYY15 pKa = 7.52TEE17 pKa = 4.32EE18 pKa = 4.27EE19 pKa = 4.09RR20 pKa = 11.84DD21 pKa = 3.48SLRR24 pKa = 11.84SLKK27 pKa = 10.3CAYY30 pKa = 9.82QVFGYY35 pKa = 10.34EE36 pKa = 3.98RR37 pKa = 11.84GAADD41 pKa = 3.75TPHH44 pKa = 6.32LQGYY48 pKa = 7.38VQFAHH53 pKa = 6.15QKK55 pKa = 8.0TLSAVKK61 pKa = 10.22KK62 pKa = 9.18LLPRR66 pKa = 11.84AHH68 pKa = 6.81LEE70 pKa = 3.97EE71 pKa = 4.21RR72 pKa = 11.84RR73 pKa = 11.84GTIDD77 pKa = 4.26QAVEE81 pKa = 3.96YY82 pKa = 9.63CKK84 pKa = 10.52KK85 pKa = 10.79DD86 pKa = 3.19GDD88 pKa = 3.81FEE90 pKa = 4.99EE91 pKa = 4.82YY92 pKa = 10.34GKK94 pKa = 11.05KK95 pKa = 10.16PMSQKK100 pKa = 10.71EE101 pKa = 3.93KK102 pKa = 10.88GKK104 pKa = 9.76EE105 pKa = 3.71EE106 pKa = 3.87KK107 pKa = 10.55NRR109 pKa = 11.84WKK111 pKa = 10.7RR112 pKa = 11.84ILEE115 pKa = 4.21KK116 pKa = 10.47ADD118 pKa = 3.58EE119 pKa = 4.81GDD121 pKa = 3.93EE122 pKa = 3.88EE123 pKa = 4.33WLRR126 pKa = 11.84EE127 pKa = 4.02NEE129 pKa = 3.8PNVAFKK135 pKa = 10.97HH136 pKa = 4.83MATFRR141 pKa = 11.84SHH143 pKa = 6.76KK144 pKa = 9.34KK145 pKa = 9.42PRR147 pKa = 11.84VGTLQYY153 pKa = 11.2EE154 pKa = 4.54EE155 pKa = 5.3TPHH158 pKa = 5.8EE159 pKa = 4.37WWVGPTGTGKK169 pKa = 9.97SRR171 pKa = 11.84KK172 pKa = 8.83AHH174 pKa = 5.54EE175 pKa = 4.93EE176 pKa = 4.26YY177 pKa = 9.97PNHH180 pKa = 6.13YY181 pKa = 10.49AKK183 pKa = 10.6EE184 pKa = 4.11KK185 pKa = 9.03NKK187 pKa = 8.6WWCGYY192 pKa = 8.35TGQEE196 pKa = 3.87TVIIEE201 pKa = 4.12EE202 pKa = 4.5ADD204 pKa = 3.7PKK206 pKa = 10.71TMEE209 pKa = 4.43HH210 pKa = 6.23LAARR214 pKa = 11.84LKK216 pKa = 10.41VWADD220 pKa = 3.92RR221 pKa = 11.84YY222 pKa = 9.85PFPGEE227 pKa = 3.63IKK229 pKa = 10.02GGRR232 pKa = 11.84IEE234 pKa = 4.69GIRR237 pKa = 11.84PLRR240 pKa = 11.84VIVISNYY247 pKa = 9.56TIEE250 pKa = 4.12EE251 pKa = 4.25CFANQNDD258 pKa = 3.94VEE260 pKa = 4.16PLRR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84FKK267 pKa = 9.93EE268 pKa = 3.96VKK270 pKa = 9.55FGEE273 pKa = 4.33RR274 pKa = 11.84TMPSPWHH281 pKa = 6.22PSYY284 pKa = 10.29TLL286 pKa = 3.27
Molecular weight: 33.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
286 |
286 |
286 |
286.0 |
33.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.294 ± 0.0 | 1.748 ± 0.0 |
3.497 ± 0.0 | 12.238 ± 0.0 |
3.147 ± 0.0 | 6.993 ± 0.0 |
3.497 ± 0.0 | 3.497 ± 0.0 |
9.79 ± 0.0 | 5.594 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.748 ± 0.0 | 3.846 ± 0.0 |
5.594 ± 0.0 | 3.147 ± 0.0 |
7.692 ± 0.0 | 3.497 ± 0.0 |
5.594 ± 0.0 | 4.545 ± 0.0 |
3.147 ± 0.0 | 4.895 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
