
Wencheng Sm shrew coronavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; unclassified Alphacoronavirus
Average proteome isoelectric point is 7.3
Get precalculated fractions of proteins
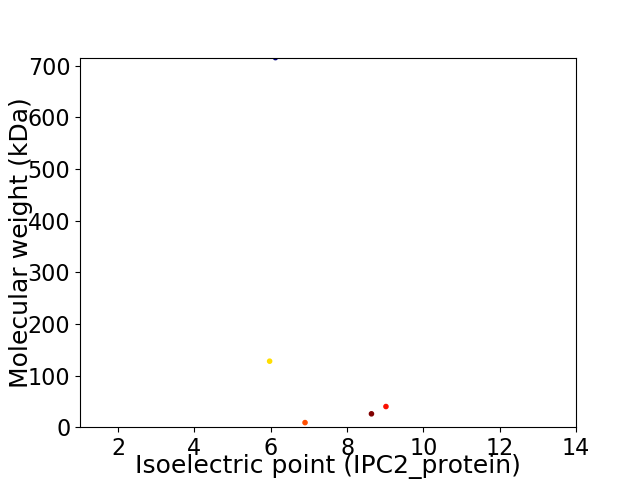
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
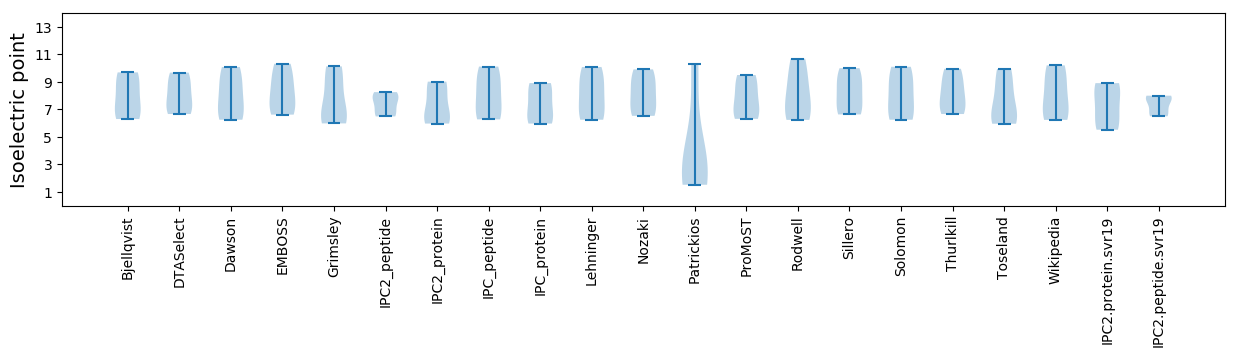
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A240FW19|A0A240FW19_9ALPC Membrane protein OS=Wencheng Sm shrew coronavirus OX=1508228 GN=M PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 10.58LIILFLVLSCVEE14 pKa = 3.78CVEE17 pKa = 4.65YY18 pKa = 10.4IKK20 pKa = 10.96CSNTQLGLLDD30 pKa = 4.44LEE32 pKa = 5.07KK33 pKa = 9.85ITYY36 pKa = 10.48SLFDD40 pKa = 4.3LGDD43 pKa = 3.4VGVNSTTLINGYY55 pKa = 9.57FPSKK59 pKa = 10.65DD60 pKa = 3.17INNWLCEE67 pKa = 3.91KK68 pKa = 10.15MGVSEE73 pKa = 5.16LYY75 pKa = 10.43YY76 pKa = 10.77GQAFYY81 pKa = 10.44TILNNVGSASLKK93 pKa = 10.57FLIQPIVYY101 pKa = 9.7SNSSYY106 pKa = 10.78VIYY109 pKa = 9.36MAYY112 pKa = 9.78KK113 pKa = 9.98SGVLNFSICYY123 pKa = 9.39RR124 pKa = 11.84NYY126 pKa = 10.39FSFSWDD132 pKa = 3.45TDD134 pKa = 3.59PPSPISCAFNRR145 pKa = 11.84RR146 pKa = 11.84IYY148 pKa = 10.38FGHH151 pKa = 6.8RR152 pKa = 11.84YY153 pKa = 9.3IYY155 pKa = 10.55LGVIWRR161 pKa = 11.84HH162 pKa = 5.38GYY164 pKa = 10.08VKK166 pKa = 10.35IITPNFVQQFEE177 pKa = 4.74LDD179 pKa = 3.45NMYY182 pKa = 11.05LEE184 pKa = 4.98VIRR187 pKa = 11.84VATGRR192 pKa = 11.84FGSTVNYY199 pKa = 9.04VIVMPFQIVDD209 pKa = 3.24KK210 pKa = 10.87RR211 pKa = 11.84STFLVNYY218 pKa = 7.53NQSGYY223 pKa = 10.78VSNLLNCSEE232 pKa = 4.78SIEE235 pKa = 4.14AQLKK239 pKa = 8.85CKK241 pKa = 10.28SRR243 pKa = 11.84SFEE246 pKa = 4.18LQSGVYY252 pKa = 9.75EE253 pKa = 3.96IALEE257 pKa = 4.02EE258 pKa = 4.13RR259 pKa = 11.84VGGYY263 pKa = 10.17YY264 pKa = 10.4SVVTNVTGDD273 pKa = 3.76CNNIKK278 pKa = 10.42HH279 pKa = 6.2LFSNFTGNRR288 pKa = 11.84QPLYY292 pKa = 10.07AGNFASMSCIVPFNFSINNDD312 pKa = 3.64LLNNNPQLRR321 pKa = 11.84GFNGEE326 pKa = 4.17FKK328 pKa = 10.85PFIDD332 pKa = 4.44CKK334 pKa = 10.59GINPIGLFGSNGTCYY349 pKa = 10.7SGMEE353 pKa = 3.51WRR355 pKa = 11.84YY356 pKa = 3.85WTRR359 pKa = 11.84RR360 pKa = 11.84FSGYY364 pKa = 10.89NMSIEE369 pKa = 4.71GIAEE373 pKa = 3.96SFYY376 pKa = 11.27DD377 pKa = 3.74AGIGEE382 pKa = 4.27NEE384 pKa = 4.65CYY386 pKa = 10.67GLLKK390 pKa = 10.29GRR392 pKa = 11.84QYY394 pKa = 11.66NEE396 pKa = 3.45FWQFTEE402 pKa = 3.61VLYY405 pKa = 10.7YY406 pKa = 10.26KK407 pKa = 10.54VRR409 pKa = 11.84FMEE412 pKa = 4.93KK413 pKa = 10.46GLDD416 pKa = 3.41VCYY419 pKa = 10.95GNLPPAPQVNKK430 pKa = 10.32CNVWTFNDD438 pKa = 3.13ITFEE442 pKa = 4.18GVLLEE447 pKa = 4.28TNKK450 pKa = 9.29TFNPQLKK457 pKa = 9.64ALYY460 pKa = 8.51KK461 pKa = 10.09GSMVVMIRR469 pKa = 11.84VFGIVYY475 pKa = 9.41EE476 pKa = 4.03VVPCIKK482 pKa = 9.7TRR484 pKa = 11.84LSVIYY489 pKa = 10.14NANNKK494 pKa = 9.68NYY496 pKa = 7.97STLYY500 pKa = 10.25RR501 pKa = 11.84VANCNQIDD509 pKa = 4.24TKK511 pKa = 10.96RR512 pKa = 11.84SVHH515 pKa = 5.6PVGLLSRR522 pKa = 11.84DD523 pKa = 4.29GINTPCGCLRR533 pKa = 11.84NAMFSPNITVDD544 pKa = 3.33SYY546 pKa = 11.71SCKK549 pKa = 9.76TLLSDD554 pKa = 4.54GYY556 pKa = 11.17CVVFNNGSTEE566 pKa = 3.98VVPIGIVGANTTYY579 pKa = 11.35NMPILEE585 pKa = 4.15EE586 pKa = 4.35SFVEE590 pKa = 4.08LALDD594 pKa = 3.36HH595 pKa = 6.45VLVSEE600 pKa = 4.37SEE602 pKa = 4.35YY603 pKa = 10.71FQTEE607 pKa = 4.13MPVFDD612 pKa = 5.1IDD614 pKa = 3.84CEE616 pKa = 4.62KK617 pKa = 10.92YY618 pKa = 10.04ICDD621 pKa = 3.51NSEE624 pKa = 3.87SCRR627 pKa = 11.84MLLEE631 pKa = 4.19QYY633 pKa = 11.17AGFCSKK639 pKa = 9.12ITSDD643 pKa = 3.94VKK645 pKa = 11.31GNSQLLNANTLAMYY659 pKa = 10.66KK660 pKa = 10.4NIVPSDD666 pKa = 3.84SIKK669 pKa = 10.51PIEE672 pKa = 4.31NFGDD676 pKa = 3.92FNMSMFIDD684 pKa = 3.44KK685 pKa = 10.48SGRR688 pKa = 11.84SVLEE692 pKa = 4.02DD693 pKa = 3.53LLFDD697 pKa = 5.27KK698 pKa = 10.72IVTTGPGFYY707 pKa = 10.02EE708 pKa = 4.98DD709 pKa = 4.46YY710 pKa = 10.12YY711 pKa = 11.16KK712 pKa = 11.06CRR714 pKa = 11.84TTTVKK719 pKa = 10.8DD720 pKa = 3.65LVCAQYY726 pKa = 11.39YY727 pKa = 8.4NGIMIIPPIQDD738 pKa = 3.12AEE740 pKa = 4.49TIGMYY745 pKa = 10.51GGIVAAGMTMGLFGGQATMGTWQIAMSARR774 pKa = 11.84LNAVGVTQGMIMEE787 pKa = 5.23DD788 pKa = 3.64VQKK791 pKa = 10.72LANGFNTLATSVSKK805 pKa = 9.93MASVTADD812 pKa = 3.17ALSRR816 pKa = 11.84IQGVVNANALQVEE829 pKa = 4.62LLARR833 pKa = 11.84ATMDD837 pKa = 3.08NFGAISSNFEE847 pKa = 4.07IIEE850 pKa = 4.04KK851 pKa = 10.31RR852 pKa = 11.84LKK854 pKa = 9.94QVQAEE859 pKa = 4.09QQMDD863 pKa = 3.53RR864 pKa = 11.84VINGKK869 pKa = 8.63MNALLNFVSNYY880 pKa = 8.58KK881 pKa = 10.57LKK883 pKa = 10.72VSEE886 pKa = 4.3LTSIQQLVKK895 pKa = 10.8QKK897 pKa = 10.19VQEE900 pKa = 4.32CVHH903 pKa = 5.87AQSLRR908 pKa = 11.84NGFCGSGLHH917 pKa = 6.54VMSIPQLVPNGVMFLHH933 pKa = 7.11FSLVKK938 pKa = 10.26NNSIIVKK945 pKa = 7.12QTPGLCKK952 pKa = 9.73EE953 pKa = 4.58DD954 pKa = 4.44KK955 pKa = 10.48INCIVPKK962 pKa = 10.57SGIFVSKK969 pKa = 10.77NDD971 pKa = 3.83SNWYY975 pKa = 5.94ITPRR979 pKa = 11.84NKK981 pKa = 9.99YY982 pKa = 8.62VPKK985 pKa = 10.6NITNSDD991 pKa = 4.04IIFTTYY997 pKa = 10.73SEE999 pKa = 4.36NFTVINNTIQFPGIEE1014 pKa = 4.38FPQDD1018 pKa = 2.77GDD1020 pKa = 3.41FDD1022 pKa = 4.2GQIKK1026 pKa = 10.54NVTLEE1031 pKa = 4.07LQSLKK1036 pKa = 11.09DD1037 pKa = 3.43VVANMSYY1044 pKa = 11.41LDD1046 pKa = 4.25LSNHH1050 pKa = 5.2TALLKK1055 pKa = 10.88NISEE1059 pKa = 4.58EE1060 pKa = 4.19VKK1062 pKa = 10.74KK1063 pKa = 10.82MNITVSQFKK1072 pKa = 10.68QYY1074 pKa = 10.53VKK1076 pKa = 9.85YY1077 pKa = 10.88VKK1079 pKa = 9.66WPWYY1083 pKa = 8.28VWLLIVMALIAFGLVCFWIFMCTGCCGLCDD1113 pKa = 4.87CIRR1116 pKa = 11.84RR1117 pKa = 11.84SCCDD1121 pKa = 3.14SNTYY1125 pKa = 10.54EE1126 pKa = 4.81PIEE1129 pKa = 4.81KK1130 pKa = 9.87IHH1132 pKa = 5.99IQQ1134 pKa = 3.28
MM1 pKa = 7.45KK2 pKa = 10.58LIILFLVLSCVEE14 pKa = 3.78CVEE17 pKa = 4.65YY18 pKa = 10.4IKK20 pKa = 10.96CSNTQLGLLDD30 pKa = 4.44LEE32 pKa = 5.07KK33 pKa = 9.85ITYY36 pKa = 10.48SLFDD40 pKa = 4.3LGDD43 pKa = 3.4VGVNSTTLINGYY55 pKa = 9.57FPSKK59 pKa = 10.65DD60 pKa = 3.17INNWLCEE67 pKa = 3.91KK68 pKa = 10.15MGVSEE73 pKa = 5.16LYY75 pKa = 10.43YY76 pKa = 10.77GQAFYY81 pKa = 10.44TILNNVGSASLKK93 pKa = 10.57FLIQPIVYY101 pKa = 9.7SNSSYY106 pKa = 10.78VIYY109 pKa = 9.36MAYY112 pKa = 9.78KK113 pKa = 9.98SGVLNFSICYY123 pKa = 9.39RR124 pKa = 11.84NYY126 pKa = 10.39FSFSWDD132 pKa = 3.45TDD134 pKa = 3.59PPSPISCAFNRR145 pKa = 11.84RR146 pKa = 11.84IYY148 pKa = 10.38FGHH151 pKa = 6.8RR152 pKa = 11.84YY153 pKa = 9.3IYY155 pKa = 10.55LGVIWRR161 pKa = 11.84HH162 pKa = 5.38GYY164 pKa = 10.08VKK166 pKa = 10.35IITPNFVQQFEE177 pKa = 4.74LDD179 pKa = 3.45NMYY182 pKa = 11.05LEE184 pKa = 4.98VIRR187 pKa = 11.84VATGRR192 pKa = 11.84FGSTVNYY199 pKa = 9.04VIVMPFQIVDD209 pKa = 3.24KK210 pKa = 10.87RR211 pKa = 11.84STFLVNYY218 pKa = 7.53NQSGYY223 pKa = 10.78VSNLLNCSEE232 pKa = 4.78SIEE235 pKa = 4.14AQLKK239 pKa = 8.85CKK241 pKa = 10.28SRR243 pKa = 11.84SFEE246 pKa = 4.18LQSGVYY252 pKa = 9.75EE253 pKa = 3.96IALEE257 pKa = 4.02EE258 pKa = 4.13RR259 pKa = 11.84VGGYY263 pKa = 10.17YY264 pKa = 10.4SVVTNVTGDD273 pKa = 3.76CNNIKK278 pKa = 10.42HH279 pKa = 6.2LFSNFTGNRR288 pKa = 11.84QPLYY292 pKa = 10.07AGNFASMSCIVPFNFSINNDD312 pKa = 3.64LLNNNPQLRR321 pKa = 11.84GFNGEE326 pKa = 4.17FKK328 pKa = 10.85PFIDD332 pKa = 4.44CKK334 pKa = 10.59GINPIGLFGSNGTCYY349 pKa = 10.7SGMEE353 pKa = 3.51WRR355 pKa = 11.84YY356 pKa = 3.85WTRR359 pKa = 11.84RR360 pKa = 11.84FSGYY364 pKa = 10.89NMSIEE369 pKa = 4.71GIAEE373 pKa = 3.96SFYY376 pKa = 11.27DD377 pKa = 3.74AGIGEE382 pKa = 4.27NEE384 pKa = 4.65CYY386 pKa = 10.67GLLKK390 pKa = 10.29GRR392 pKa = 11.84QYY394 pKa = 11.66NEE396 pKa = 3.45FWQFTEE402 pKa = 3.61VLYY405 pKa = 10.7YY406 pKa = 10.26KK407 pKa = 10.54VRR409 pKa = 11.84FMEE412 pKa = 4.93KK413 pKa = 10.46GLDD416 pKa = 3.41VCYY419 pKa = 10.95GNLPPAPQVNKK430 pKa = 10.32CNVWTFNDD438 pKa = 3.13ITFEE442 pKa = 4.18GVLLEE447 pKa = 4.28TNKK450 pKa = 9.29TFNPQLKK457 pKa = 9.64ALYY460 pKa = 8.51KK461 pKa = 10.09GSMVVMIRR469 pKa = 11.84VFGIVYY475 pKa = 9.41EE476 pKa = 4.03VVPCIKK482 pKa = 9.7TRR484 pKa = 11.84LSVIYY489 pKa = 10.14NANNKK494 pKa = 9.68NYY496 pKa = 7.97STLYY500 pKa = 10.25RR501 pKa = 11.84VANCNQIDD509 pKa = 4.24TKK511 pKa = 10.96RR512 pKa = 11.84SVHH515 pKa = 5.6PVGLLSRR522 pKa = 11.84DD523 pKa = 4.29GINTPCGCLRR533 pKa = 11.84NAMFSPNITVDD544 pKa = 3.33SYY546 pKa = 11.71SCKK549 pKa = 9.76TLLSDD554 pKa = 4.54GYY556 pKa = 11.17CVVFNNGSTEE566 pKa = 3.98VVPIGIVGANTTYY579 pKa = 11.35NMPILEE585 pKa = 4.15EE586 pKa = 4.35SFVEE590 pKa = 4.08LALDD594 pKa = 3.36HH595 pKa = 6.45VLVSEE600 pKa = 4.37SEE602 pKa = 4.35YY603 pKa = 10.71FQTEE607 pKa = 4.13MPVFDD612 pKa = 5.1IDD614 pKa = 3.84CEE616 pKa = 4.62KK617 pKa = 10.92YY618 pKa = 10.04ICDD621 pKa = 3.51NSEE624 pKa = 3.87SCRR627 pKa = 11.84MLLEE631 pKa = 4.19QYY633 pKa = 11.17AGFCSKK639 pKa = 9.12ITSDD643 pKa = 3.94VKK645 pKa = 11.31GNSQLLNANTLAMYY659 pKa = 10.66KK660 pKa = 10.4NIVPSDD666 pKa = 3.84SIKK669 pKa = 10.51PIEE672 pKa = 4.31NFGDD676 pKa = 3.92FNMSMFIDD684 pKa = 3.44KK685 pKa = 10.48SGRR688 pKa = 11.84SVLEE692 pKa = 4.02DD693 pKa = 3.53LLFDD697 pKa = 5.27KK698 pKa = 10.72IVTTGPGFYY707 pKa = 10.02EE708 pKa = 4.98DD709 pKa = 4.46YY710 pKa = 10.12YY711 pKa = 11.16KK712 pKa = 11.06CRR714 pKa = 11.84TTTVKK719 pKa = 10.8DD720 pKa = 3.65LVCAQYY726 pKa = 11.39YY727 pKa = 8.4NGIMIIPPIQDD738 pKa = 3.12AEE740 pKa = 4.49TIGMYY745 pKa = 10.51GGIVAAGMTMGLFGGQATMGTWQIAMSARR774 pKa = 11.84LNAVGVTQGMIMEE787 pKa = 5.23DD788 pKa = 3.64VQKK791 pKa = 10.72LANGFNTLATSVSKK805 pKa = 9.93MASVTADD812 pKa = 3.17ALSRR816 pKa = 11.84IQGVVNANALQVEE829 pKa = 4.62LLARR833 pKa = 11.84ATMDD837 pKa = 3.08NFGAISSNFEE847 pKa = 4.07IIEE850 pKa = 4.04KK851 pKa = 10.31RR852 pKa = 11.84LKK854 pKa = 9.94QVQAEE859 pKa = 4.09QQMDD863 pKa = 3.53RR864 pKa = 11.84VINGKK869 pKa = 8.63MNALLNFVSNYY880 pKa = 8.58KK881 pKa = 10.57LKK883 pKa = 10.72VSEE886 pKa = 4.3LTSIQQLVKK895 pKa = 10.8QKK897 pKa = 10.19VQEE900 pKa = 4.32CVHH903 pKa = 5.87AQSLRR908 pKa = 11.84NGFCGSGLHH917 pKa = 6.54VMSIPQLVPNGVMFLHH933 pKa = 7.11FSLVKK938 pKa = 10.26NNSIIVKK945 pKa = 7.12QTPGLCKK952 pKa = 9.73EE953 pKa = 4.58DD954 pKa = 4.44KK955 pKa = 10.48INCIVPKK962 pKa = 10.57SGIFVSKK969 pKa = 10.77NDD971 pKa = 3.83SNWYY975 pKa = 5.94ITPRR979 pKa = 11.84NKK981 pKa = 9.99YY982 pKa = 8.62VPKK985 pKa = 10.6NITNSDD991 pKa = 4.04IIFTTYY997 pKa = 10.73SEE999 pKa = 4.36NFTVINNTIQFPGIEE1014 pKa = 4.38FPQDD1018 pKa = 2.77GDD1020 pKa = 3.41FDD1022 pKa = 4.2GQIKK1026 pKa = 10.54NVTLEE1031 pKa = 4.07LQSLKK1036 pKa = 11.09DD1037 pKa = 3.43VVANMSYY1044 pKa = 11.41LDD1046 pKa = 4.25LSNHH1050 pKa = 5.2TALLKK1055 pKa = 10.88NISEE1059 pKa = 4.58EE1060 pKa = 4.19VKK1062 pKa = 10.74KK1063 pKa = 10.82MNITVSQFKK1072 pKa = 10.68QYY1074 pKa = 10.53VKK1076 pKa = 9.85YY1077 pKa = 10.88VKK1079 pKa = 9.66WPWYY1083 pKa = 8.28VWLLIVMALIAFGLVCFWIFMCTGCCGLCDD1113 pKa = 4.87CIRR1116 pKa = 11.84RR1117 pKa = 11.84SCCDD1121 pKa = 3.14SNTYY1125 pKa = 10.54EE1126 pKa = 4.81PIEE1129 pKa = 4.81KK1130 pKa = 9.87IHH1132 pKa = 5.99IQQ1134 pKa = 3.28
Molecular weight: 127.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A240FW20|A0A240FW20_9ALPC Nucleocapsid protein OS=Wencheng Sm shrew coronavirus OX=1508228 PE=4 SV=1
MM1 pKa = 7.21SAEE4 pKa = 4.18KK5 pKa = 10.79VSWSSSLPTKK15 pKa = 10.76ADD17 pKa = 3.29TKK19 pKa = 10.6NGSKK23 pKa = 10.57VEE25 pKa = 4.19VEE27 pKa = 4.21LSAWDD32 pKa = 4.38PIIIQNNKK40 pKa = 9.49NIQDD44 pKa = 3.75VLPRR48 pKa = 11.84TSVPKK53 pKa = 10.36GLPSGHH59 pKa = 6.89EE60 pKa = 3.01IGYY63 pKa = 8.73WYY65 pKa = 10.25KK66 pKa = 10.57GPEE69 pKa = 3.71RR70 pKa = 11.84YY71 pKa = 9.73RR72 pKa = 11.84FTRR75 pKa = 11.84GEE77 pKa = 4.25KK78 pKa = 10.68VPLDD82 pKa = 4.07PKK84 pKa = 10.2WYY86 pKa = 9.62FYY88 pKa = 10.77PLGHH92 pKa = 6.72GPAANKK98 pKa = 9.38KK99 pKa = 8.03WNSKK103 pKa = 9.7VEE105 pKa = 4.15GVFWVARR112 pKa = 11.84DD113 pKa = 3.43GASIEE118 pKa = 4.15KK119 pKa = 10.67VPDD122 pKa = 3.65FEE124 pKa = 4.43PRR126 pKa = 11.84KK127 pKa = 10.13KK128 pKa = 10.22GAEE131 pKa = 4.02AKK133 pKa = 9.89RR134 pKa = 11.84VRR136 pKa = 11.84LPGKK140 pKa = 10.31APANVYY146 pKa = 7.6FTTTSASRR154 pKa = 11.84SQSRR158 pKa = 11.84DD159 pKa = 2.72NSRR162 pKa = 11.84AASRR166 pKa = 11.84SGSKK170 pKa = 10.33SRR172 pKa = 11.84IQSRR176 pKa = 11.84SPSAEE181 pKa = 3.68RR182 pKa = 11.84GHH184 pKa = 8.04VDD186 pKa = 2.46IKK188 pKa = 11.11LAVLEE193 pKa = 4.05ALKK196 pKa = 10.87DD197 pKa = 3.66LGIGQDD203 pKa = 3.53TKK205 pKa = 11.27KK206 pKa = 10.55KK207 pKa = 9.22SSKK210 pKa = 10.21SAAASGNNTPKK221 pKa = 10.22QEE223 pKa = 4.34SSPKK227 pKa = 8.63VQRR230 pKa = 11.84KK231 pKa = 7.91QIEE234 pKa = 4.19RR235 pKa = 11.84LRR237 pKa = 11.84HH238 pKa = 4.91KK239 pKa = 10.31RR240 pKa = 11.84VPTGKK245 pKa = 10.41EE246 pKa = 3.68NITVCFGPRR255 pKa = 11.84GPEE258 pKa = 3.58QNFGSDD264 pKa = 3.54EE265 pKa = 4.12IVNGGAQGKK274 pKa = 9.16KK275 pKa = 8.73VAQLLEE281 pKa = 4.29NVPGPSALLFGGKK294 pKa = 9.2VEE296 pKa = 5.11QIGKK300 pKa = 10.29ANGKK304 pKa = 9.24VAVQYY309 pKa = 10.42TYY311 pKa = 11.2VAEE314 pKa = 4.57LDD316 pKa = 3.84DD317 pKa = 4.2NEE319 pKa = 4.27EE320 pKa = 4.01VKK322 pKa = 10.82RR323 pKa = 11.84FLDD326 pKa = 5.62LIDD329 pKa = 4.85AYY331 pKa = 10.01KK332 pKa = 10.69KK333 pKa = 8.49PTTANSINSVLNPQATAFAPPSGVIEE359 pKa = 4.17EE360 pKa = 4.77LQDD363 pKa = 4.05DD364 pKa = 4.5LNN366 pKa = 4.44
MM1 pKa = 7.21SAEE4 pKa = 4.18KK5 pKa = 10.79VSWSSSLPTKK15 pKa = 10.76ADD17 pKa = 3.29TKK19 pKa = 10.6NGSKK23 pKa = 10.57VEE25 pKa = 4.19VEE27 pKa = 4.21LSAWDD32 pKa = 4.38PIIIQNNKK40 pKa = 9.49NIQDD44 pKa = 3.75VLPRR48 pKa = 11.84TSVPKK53 pKa = 10.36GLPSGHH59 pKa = 6.89EE60 pKa = 3.01IGYY63 pKa = 8.73WYY65 pKa = 10.25KK66 pKa = 10.57GPEE69 pKa = 3.71RR70 pKa = 11.84YY71 pKa = 9.73RR72 pKa = 11.84FTRR75 pKa = 11.84GEE77 pKa = 4.25KK78 pKa = 10.68VPLDD82 pKa = 4.07PKK84 pKa = 10.2WYY86 pKa = 9.62FYY88 pKa = 10.77PLGHH92 pKa = 6.72GPAANKK98 pKa = 9.38KK99 pKa = 8.03WNSKK103 pKa = 9.7VEE105 pKa = 4.15GVFWVARR112 pKa = 11.84DD113 pKa = 3.43GASIEE118 pKa = 4.15KK119 pKa = 10.67VPDD122 pKa = 3.65FEE124 pKa = 4.43PRR126 pKa = 11.84KK127 pKa = 10.13KK128 pKa = 10.22GAEE131 pKa = 4.02AKK133 pKa = 9.89RR134 pKa = 11.84VRR136 pKa = 11.84LPGKK140 pKa = 10.31APANVYY146 pKa = 7.6FTTTSASRR154 pKa = 11.84SQSRR158 pKa = 11.84DD159 pKa = 2.72NSRR162 pKa = 11.84AASRR166 pKa = 11.84SGSKK170 pKa = 10.33SRR172 pKa = 11.84IQSRR176 pKa = 11.84SPSAEE181 pKa = 3.68RR182 pKa = 11.84GHH184 pKa = 8.04VDD186 pKa = 2.46IKK188 pKa = 11.11LAVLEE193 pKa = 4.05ALKK196 pKa = 10.87DD197 pKa = 3.66LGIGQDD203 pKa = 3.53TKK205 pKa = 11.27KK206 pKa = 10.55KK207 pKa = 9.22SSKK210 pKa = 10.21SAAASGNNTPKK221 pKa = 10.22QEE223 pKa = 4.34SSPKK227 pKa = 8.63VQRR230 pKa = 11.84KK231 pKa = 7.91QIEE234 pKa = 4.19RR235 pKa = 11.84LRR237 pKa = 11.84HH238 pKa = 4.91KK239 pKa = 10.31RR240 pKa = 11.84VPTGKK245 pKa = 10.41EE246 pKa = 3.68NITVCFGPRR255 pKa = 11.84GPEE258 pKa = 3.58QNFGSDD264 pKa = 3.54EE265 pKa = 4.12IVNGGAQGKK274 pKa = 9.16KK275 pKa = 8.73VAQLLEE281 pKa = 4.29NVPGPSALLFGGKK294 pKa = 9.2VEE296 pKa = 5.11QIGKK300 pKa = 10.29ANGKK304 pKa = 9.24VAVQYY309 pKa = 10.42TYY311 pKa = 11.2VAEE314 pKa = 4.57LDD316 pKa = 3.84DD317 pKa = 4.2NEE319 pKa = 4.27EE320 pKa = 4.01VKK322 pKa = 10.82RR323 pKa = 11.84FLDD326 pKa = 5.62LIDD329 pKa = 4.85AYY331 pKa = 10.01KK332 pKa = 10.69KK333 pKa = 8.49PTTANSINSVLNPQATAFAPPSGVIEE359 pKa = 4.17EE360 pKa = 4.77LQDD363 pKa = 4.05DD364 pKa = 4.5LNN366 pKa = 4.44
Molecular weight: 40.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8128 |
75 |
6324 |
1625.6 |
183.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.798 ± 0.75 | 3.088 ± 0.665 |
5.376 ± 0.869 | 4.86 ± 0.415 |
6.115 ± 0.731 | 6.299 ± 0.424 |
1.513 ± 0.255 | 5.598 ± 0.831 |
7.16 ± 0.891 | 8.981 ± 0.715 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.387 ± 0.49 | 7.013 ± 0.574 |
2.879 ± 1.123 | 2.842 ± 0.479 |
3.027 ± 0.67 | 7.013 ± 0.665 |
4.737 ± 0.325 | 10.162 ± 0.954 |
1.28 ± 0.276 | 4.872 ± 0.488 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
