
West African Asystasia virus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 7.8
Get precalculated fractions of proteins
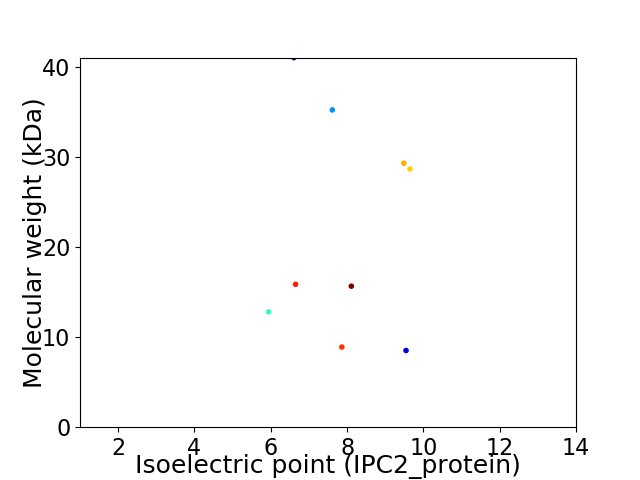
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A190D9H7|A0A190D9H7_9GEMI AC5 OS=West African Asystasia virus 1 OX=1046573 PE=4 SV=1
MM1 pKa = 8.12WDD3 pKa = 3.43PLLNSFPPTIHH14 pKa = 7.19GFRR17 pKa = 11.84CMLALKK23 pKa = 10.45YY24 pKa = 10.82LLLLEE29 pKa = 4.91NNYY32 pKa = 10.2EE33 pKa = 4.22DD34 pKa = 4.24NSVGQVYY41 pKa = 9.85IRR43 pKa = 11.84EE44 pKa = 4.56LISVLRR50 pKa = 11.84AGDD53 pKa = 3.67YY54 pKa = 10.72VKK56 pKa = 10.91ASSRR60 pKa = 11.84YY61 pKa = 7.7CDD63 pKa = 4.2LYY65 pKa = 10.75PRR67 pKa = 11.84IQGTSPPEE75 pKa = 3.64LRR77 pKa = 11.84QPSCQCTKK85 pKa = 10.48CPRR88 pKa = 11.84HH89 pKa = 5.69QKK91 pKa = 10.77EE92 pKa = 4.38SMGEE96 pKa = 3.84QAHH99 pKa = 5.45VSEE102 pKa = 4.74TSSVPEE108 pKa = 4.4VYY110 pKa = 10.25QSS112 pKa = 2.97
MM1 pKa = 8.12WDD3 pKa = 3.43PLLNSFPPTIHH14 pKa = 7.19GFRR17 pKa = 11.84CMLALKK23 pKa = 10.45YY24 pKa = 10.82LLLLEE29 pKa = 4.91NNYY32 pKa = 10.2EE33 pKa = 4.22DD34 pKa = 4.24NSVGQVYY41 pKa = 9.85IRR43 pKa = 11.84EE44 pKa = 4.56LISVLRR50 pKa = 11.84AGDD53 pKa = 3.67YY54 pKa = 10.72VKK56 pKa = 10.91ASSRR60 pKa = 11.84YY61 pKa = 7.7CDD63 pKa = 4.2LYY65 pKa = 10.75PRR67 pKa = 11.84IQGTSPPEE75 pKa = 3.64LRR77 pKa = 11.84QPSCQCTKK85 pKa = 10.48CPRR88 pKa = 11.84HH89 pKa = 5.69QKK91 pKa = 10.77EE92 pKa = 4.38SMGEE96 pKa = 3.84QAHH99 pKa = 5.45VSEE102 pKa = 4.74TSSVPEE108 pKa = 4.4VYY110 pKa = 10.25QSS112 pKa = 2.97
Molecular weight: 12.81 kDa
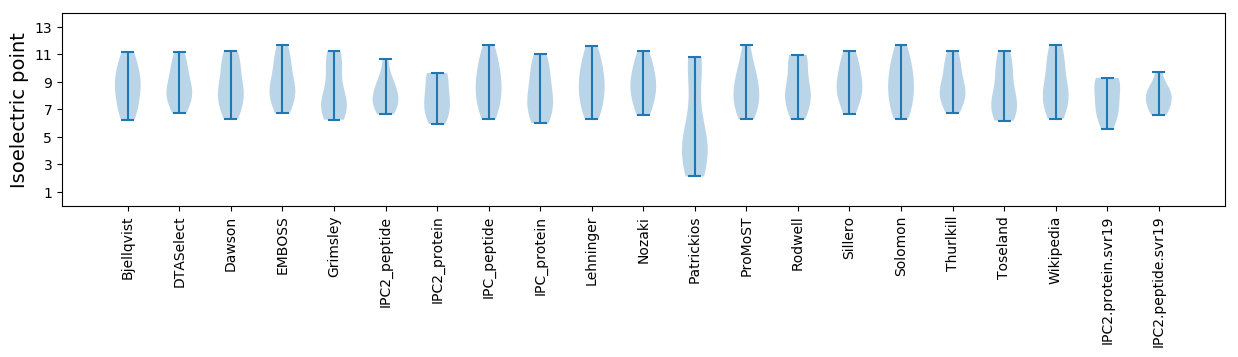
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A190D9I4|A0A190D9I4_9GEMI AC4 OS=West African Asystasia virus 1 OX=1046573 PE=3 SV=1
MM1 pKa = 7.71SKK3 pKa = 10.35RR4 pKa = 11.84PADD7 pKa = 3.52IVISTPASKK16 pKa = 10.29VRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84LNFDD24 pKa = 3.26SPRR27 pKa = 11.84ASVPNVRR34 pKa = 11.84VTRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84VWANRR44 pKa = 11.84PMYY47 pKa = 9.98RR48 pKa = 11.84KK49 pKa = 8.43PALYY53 pKa = 9.81RR54 pKa = 11.84KK55 pKa = 7.68YY56 pKa = 9.22TSRR59 pKa = 11.84DD60 pKa = 3.12VPRR63 pKa = 11.84GCEE66 pKa = 4.36GPCKK70 pKa = 10.33VQSFDD75 pKa = 3.23QRR77 pKa = 11.84DD78 pKa = 4.04DD79 pKa = 3.53IKK81 pKa = 10.95HH82 pKa = 6.1LGVVRR87 pKa = 11.84CISDD91 pKa = 3.36VTRR94 pKa = 11.84GPGITHH100 pKa = 6.76RR101 pKa = 11.84VGKK104 pKa = 9.62RR105 pKa = 11.84FCIKK109 pKa = 10.44SVLFTGKK116 pKa = 8.58IWMDD120 pKa = 3.46EE121 pKa = 4.13NIKK124 pKa = 10.05KK125 pKa = 10.0QNHH128 pKa = 4.27TNIVIFFLVRR138 pKa = 11.84DD139 pKa = 3.84RR140 pKa = 11.84RR141 pKa = 11.84PYY143 pKa = 10.9GSPQDD148 pKa = 3.87FGDD151 pKa = 3.79VFNMFDD157 pKa = 4.34NEE159 pKa = 4.11PSTATVKK166 pKa = 10.81NDD168 pKa = 2.84LRR170 pKa = 11.84DD171 pKa = 3.41RR172 pKa = 11.84YY173 pKa = 9.78QVLRR177 pKa = 11.84RR178 pKa = 11.84FTCSVTGGPSGCKK191 pKa = 8.82EE192 pKa = 3.67QALVRR197 pKa = 11.84RR198 pKa = 11.84FYY200 pKa = 10.55TINHH204 pKa = 5.37NVVYY208 pKa = 10.24NHH210 pKa = 5.98QEE212 pKa = 3.39AAKK215 pKa = 10.4YY216 pKa = 9.25EE217 pKa = 4.13NHH219 pKa = 6.46TEE221 pKa = 3.99NALLLYY227 pKa = 7.29MACTHH232 pKa = 7.07ASNPVYY238 pKa = 10.85ASMKK242 pKa = 8.99IRR244 pKa = 11.84IYY246 pKa = 10.61FYY248 pKa = 11.2DD249 pKa = 3.78SIGNN253 pKa = 3.69
MM1 pKa = 7.71SKK3 pKa = 10.35RR4 pKa = 11.84PADD7 pKa = 3.52IVISTPASKK16 pKa = 10.29VRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84LNFDD24 pKa = 3.26SPRR27 pKa = 11.84ASVPNVRR34 pKa = 11.84VTRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84VWANRR44 pKa = 11.84PMYY47 pKa = 9.98RR48 pKa = 11.84KK49 pKa = 8.43PALYY53 pKa = 9.81RR54 pKa = 11.84KK55 pKa = 7.68YY56 pKa = 9.22TSRR59 pKa = 11.84DD60 pKa = 3.12VPRR63 pKa = 11.84GCEE66 pKa = 4.36GPCKK70 pKa = 10.33VQSFDD75 pKa = 3.23QRR77 pKa = 11.84DD78 pKa = 4.04DD79 pKa = 3.53IKK81 pKa = 10.95HH82 pKa = 6.1LGVVRR87 pKa = 11.84CISDD91 pKa = 3.36VTRR94 pKa = 11.84GPGITHH100 pKa = 6.76RR101 pKa = 11.84VGKK104 pKa = 9.62RR105 pKa = 11.84FCIKK109 pKa = 10.44SVLFTGKK116 pKa = 8.58IWMDD120 pKa = 3.46EE121 pKa = 4.13NIKK124 pKa = 10.05KK125 pKa = 10.0QNHH128 pKa = 4.27TNIVIFFLVRR138 pKa = 11.84DD139 pKa = 3.84RR140 pKa = 11.84RR141 pKa = 11.84PYY143 pKa = 10.9GSPQDD148 pKa = 3.87FGDD151 pKa = 3.79VFNMFDD157 pKa = 4.34NEE159 pKa = 4.11PSTATVKK166 pKa = 10.81NDD168 pKa = 2.84LRR170 pKa = 11.84DD171 pKa = 3.41RR172 pKa = 11.84YY173 pKa = 9.78QVLRR177 pKa = 11.84RR178 pKa = 11.84FTCSVTGGPSGCKK191 pKa = 8.82EE192 pKa = 3.67QALVRR197 pKa = 11.84RR198 pKa = 11.84FYY200 pKa = 10.55TINHH204 pKa = 5.37NVVYY208 pKa = 10.24NHH210 pKa = 5.98QEE212 pKa = 3.39AAKK215 pKa = 10.4YY216 pKa = 9.25EE217 pKa = 4.13NHH219 pKa = 6.46TEE221 pKa = 3.99NALLLYY227 pKa = 7.29MACTHH232 pKa = 7.07ASNPVYY238 pKa = 10.85ASMKK242 pKa = 8.99IRR244 pKa = 11.84IYY246 pKa = 10.61FYY248 pKa = 11.2DD249 pKa = 3.78SIGNN253 pKa = 3.69
Molecular weight: 29.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1722 |
77 |
362 |
191.3 |
21.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.401 ± 0.592 | 2.091 ± 0.305 |
4.646 ± 0.382 | 4.588 ± 0.655 |
4.007 ± 0.437 | 5.168 ± 0.337 |
3.717 ± 0.618 | 6.446 ± 0.725 |
4.936 ± 0.623 | 6.794 ± 0.497 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.207 ± 0.266 | 5.459 ± 0.431 |
6.156 ± 0.859 | 4.239 ± 0.601 |
7.375 ± 1.001 | 9.466 ± 0.926 |
6.156 ± 0.831 | 5.807 ± 0.862 |
1.22 ± 0.228 | 4.123 ± 0.541 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
