
Camellia sinensis var. sinensis
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; Ericales; Theaceae; Camellia; Camellia sinensis
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
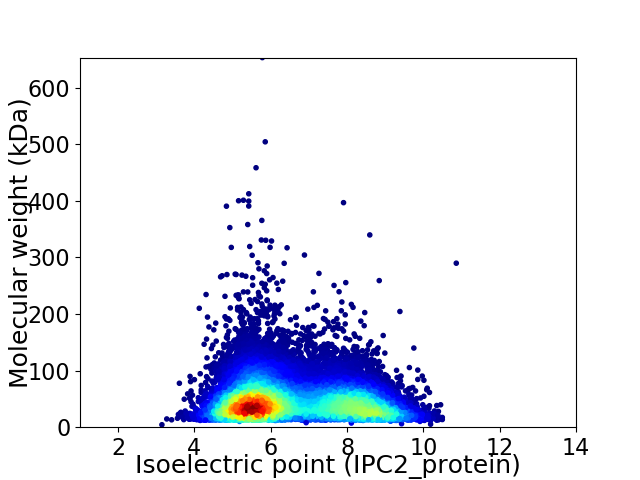
Virtual 2D-PAGE plot for 30052 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4S4DFN4|A0A4S4DFN4_CAMSI Uncharacterized protein OS=Camellia sinensis var. sinensis OX=542762 GN=TEA_020291 PE=4 SV=1
MM1 pKa = 7.75AAFLYY6 pKa = 10.48FDD8 pKa = 6.13LIINYY13 pKa = 8.65KK14 pKa = 10.54DD15 pKa = 3.25KK16 pKa = 11.09VGRR19 pKa = 11.84IYY21 pKa = 11.03DD22 pKa = 3.67VDD24 pKa = 3.99PNMYY28 pKa = 10.19CYY30 pKa = 10.13IDD32 pKa = 3.77ALKK35 pKa = 10.82DD36 pKa = 3.35VTEE39 pKa = 4.53TVLSHH44 pKa = 6.75IDD46 pKa = 3.45CSEE49 pKa = 4.48AIAITLHH56 pKa = 7.01YY57 pKa = 10.7DD58 pKa = 3.23MPGTEE63 pKa = 4.08NKK65 pKa = 10.18RR66 pKa = 11.84LIEE69 pKa = 4.28NDD71 pKa = 3.69LDD73 pKa = 3.96VLDD76 pKa = 4.32MFYY79 pKa = 10.82VQCRR83 pKa = 11.84SKK85 pKa = 9.93TINLYY90 pKa = 10.71VDD92 pKa = 3.46IAYY95 pKa = 10.48CIGKK99 pKa = 9.76EE100 pKa = 3.99DD101 pKa = 3.84GEE103 pKa = 4.48GRR105 pKa = 11.84NDD107 pKa = 3.29VGDD110 pKa = 3.85NNGGDD115 pKa = 4.0GNGGVHH121 pKa = 6.41GVHH124 pKa = 6.56TIEE127 pKa = 3.77VDD129 pKa = 3.08EE130 pKa = 5.23EE131 pKa = 4.06YY132 pKa = 10.89RR133 pKa = 11.84EE134 pKa = 4.15DD135 pKa = 3.59NVYY138 pKa = 10.82GFSDD142 pKa = 4.81EE143 pKa = 4.75DD144 pKa = 4.2RR145 pKa = 11.84DD146 pKa = 3.71WNASVDD152 pKa = 4.01GQHH155 pKa = 6.27EE156 pKa = 4.58LNGTVSLSDD165 pKa = 3.56EE166 pKa = 4.68DD167 pKa = 4.73NEE169 pKa = 5.16EE170 pKa = 4.23SNEE173 pKa = 3.89DD174 pKa = 4.29DD175 pKa = 4.68GLSDD179 pKa = 3.79YY180 pKa = 11.25QSGDD184 pKa = 3.26DD185 pKa = 3.55EE186 pKa = 4.6VRR188 pKa = 11.84YY189 pKa = 10.41SSSDD193 pKa = 3.42DD194 pKa = 3.45EE195 pKa = 4.61FAGG198 pKa = 4.09
MM1 pKa = 7.75AAFLYY6 pKa = 10.48FDD8 pKa = 6.13LIINYY13 pKa = 8.65KK14 pKa = 10.54DD15 pKa = 3.25KK16 pKa = 11.09VGRR19 pKa = 11.84IYY21 pKa = 11.03DD22 pKa = 3.67VDD24 pKa = 3.99PNMYY28 pKa = 10.19CYY30 pKa = 10.13IDD32 pKa = 3.77ALKK35 pKa = 10.82DD36 pKa = 3.35VTEE39 pKa = 4.53TVLSHH44 pKa = 6.75IDD46 pKa = 3.45CSEE49 pKa = 4.48AIAITLHH56 pKa = 7.01YY57 pKa = 10.7DD58 pKa = 3.23MPGTEE63 pKa = 4.08NKK65 pKa = 10.18RR66 pKa = 11.84LIEE69 pKa = 4.28NDD71 pKa = 3.69LDD73 pKa = 3.96VLDD76 pKa = 4.32MFYY79 pKa = 10.82VQCRR83 pKa = 11.84SKK85 pKa = 9.93TINLYY90 pKa = 10.71VDD92 pKa = 3.46IAYY95 pKa = 10.48CIGKK99 pKa = 9.76EE100 pKa = 3.99DD101 pKa = 3.84GEE103 pKa = 4.48GRR105 pKa = 11.84NDD107 pKa = 3.29VGDD110 pKa = 3.85NNGGDD115 pKa = 4.0GNGGVHH121 pKa = 6.41GVHH124 pKa = 6.56TIEE127 pKa = 3.77VDD129 pKa = 3.08EE130 pKa = 5.23EE131 pKa = 4.06YY132 pKa = 10.89RR133 pKa = 11.84EE134 pKa = 4.15DD135 pKa = 3.59NVYY138 pKa = 10.82GFSDD142 pKa = 4.81EE143 pKa = 4.75DD144 pKa = 4.2RR145 pKa = 11.84DD146 pKa = 3.71WNASVDD152 pKa = 4.01GQHH155 pKa = 6.27EE156 pKa = 4.58LNGTVSLSDD165 pKa = 3.56EE166 pKa = 4.68DD167 pKa = 4.73NEE169 pKa = 5.16EE170 pKa = 4.23SNEE173 pKa = 3.89DD174 pKa = 4.29DD175 pKa = 4.68GLSDD179 pKa = 3.79YY180 pKa = 11.25QSGDD184 pKa = 3.26DD185 pKa = 3.55EE186 pKa = 4.6VRR188 pKa = 11.84YY189 pKa = 10.41SSSDD193 pKa = 3.42DD194 pKa = 3.45EE195 pKa = 4.61FAGG198 pKa = 4.09
Molecular weight: 22.25 kDa
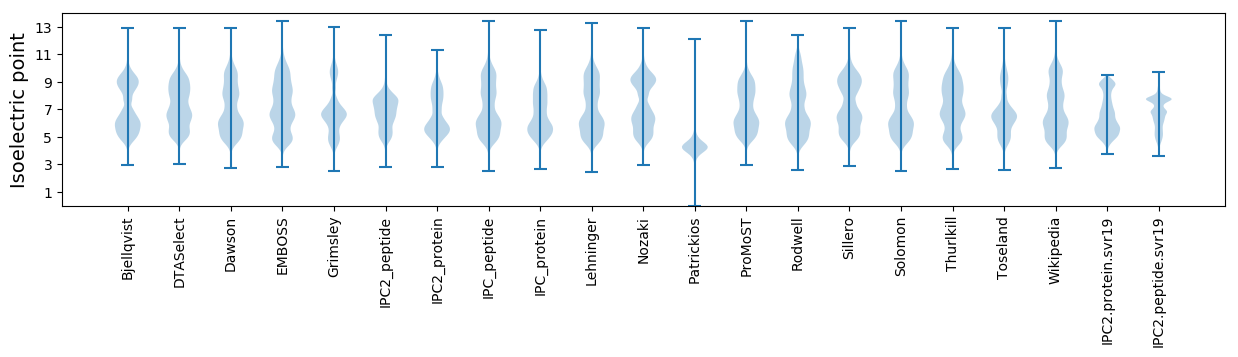
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4S4D3Q4|A0A4S4D3Q4_CAMSI Uncharacterized protein OS=Camellia sinensis var. sinensis OX=542762 GN=TEA_009666 PE=4 SV=1
MM1 pKa = 7.23SAVAKK6 pKa = 10.36ASKK9 pKa = 10.48NVFATGYY16 pKa = 7.64YY17 pKa = 8.85AHH19 pKa = 7.34RR20 pKa = 11.84RR21 pKa = 11.84STAPPFVLAAAPPQHH36 pKa = 6.15RR37 pKa = 11.84TTVRR41 pKa = 11.84PRR43 pKa = 11.84RR44 pKa = 11.84STAAAPHH51 pKa = 6.33HH52 pKa = 6.66RR53 pKa = 11.84SSSPQHH59 pKa = 6.22RR60 pKa = 11.84RR61 pKa = 11.84STAPPFVLAAAPPQHH76 pKa = 6.15RR77 pKa = 11.84TTVRR81 pKa = 11.84PRR83 pKa = 11.84RR84 pKa = 11.84STAAAPHH91 pKa = 6.33HH92 pKa = 6.66RR93 pKa = 11.84SSSPQHH99 pKa = 6.22RR100 pKa = 11.84RR101 pKa = 11.84STAPPFVLAAAPPQHH116 pKa = 6.15RR117 pKa = 11.84TTVRR121 pKa = 11.84PRR123 pKa = 11.84RR124 pKa = 11.84STAAAPHH131 pKa = 6.33HH132 pKa = 6.66RR133 pKa = 11.84SSSPQHH139 pKa = 6.22RR140 pKa = 11.84RR141 pKa = 11.84STAPPFVLAAAPPQHH156 pKa = 6.15RR157 pKa = 11.84TTVRR161 pKa = 11.84PRR163 pKa = 11.84RR164 pKa = 11.84STAAAPHH171 pKa = 6.33HH172 pKa = 6.66RR173 pKa = 11.84SSSPQHH179 pKa = 6.22RR180 pKa = 11.84RR181 pKa = 11.84STAPPFVLAAAPPQHH196 pKa = 6.15RR197 pKa = 11.84TTVRR201 pKa = 11.84PRR203 pKa = 11.84RR204 pKa = 11.84STAAAPHH211 pKa = 6.33HH212 pKa = 6.66RR213 pKa = 11.84SSSPQHH219 pKa = 6.22RR220 pKa = 11.84RR221 pKa = 11.84STAPPFVLAAAPPQHH236 pKa = 6.15RR237 pKa = 11.84TTVRR241 pKa = 11.84PRR243 pKa = 11.84RR244 pKa = 11.84STAAAPHH251 pKa = 6.33HH252 pKa = 6.66RR253 pKa = 11.84SSSPQHH259 pKa = 6.22RR260 pKa = 11.84RR261 pKa = 11.84STAPPFVLAAAPPQHH276 pKa = 6.15RR277 pKa = 11.84TTVRR281 pKa = 11.84PRR283 pKa = 11.84RR284 pKa = 11.84STAAAPHH291 pKa = 6.33HH292 pKa = 6.66RR293 pKa = 11.84SSSPQHH299 pKa = 6.22RR300 pKa = 11.84RR301 pKa = 11.84STAPPFVLAAAPPQYY316 pKa = 9.25RR317 pKa = 11.84TTVRR321 pKa = 11.84PRR323 pKa = 11.84RR324 pKa = 11.84STAAAPHH331 pKa = 6.07HH332 pKa = 6.49RR333 pKa = 11.84SPFGFAGIWGKK344 pKa = 10.67EE345 pKa = 3.54IGFTDD350 pKa = 3.79LGIWII355 pKa = 4.82
MM1 pKa = 7.23SAVAKK6 pKa = 10.36ASKK9 pKa = 10.48NVFATGYY16 pKa = 7.64YY17 pKa = 8.85AHH19 pKa = 7.34RR20 pKa = 11.84RR21 pKa = 11.84STAPPFVLAAAPPQHH36 pKa = 6.15RR37 pKa = 11.84TTVRR41 pKa = 11.84PRR43 pKa = 11.84RR44 pKa = 11.84STAAAPHH51 pKa = 6.33HH52 pKa = 6.66RR53 pKa = 11.84SSSPQHH59 pKa = 6.22RR60 pKa = 11.84RR61 pKa = 11.84STAPPFVLAAAPPQHH76 pKa = 6.15RR77 pKa = 11.84TTVRR81 pKa = 11.84PRR83 pKa = 11.84RR84 pKa = 11.84STAAAPHH91 pKa = 6.33HH92 pKa = 6.66RR93 pKa = 11.84SSSPQHH99 pKa = 6.22RR100 pKa = 11.84RR101 pKa = 11.84STAPPFVLAAAPPQHH116 pKa = 6.15RR117 pKa = 11.84TTVRR121 pKa = 11.84PRR123 pKa = 11.84RR124 pKa = 11.84STAAAPHH131 pKa = 6.33HH132 pKa = 6.66RR133 pKa = 11.84SSSPQHH139 pKa = 6.22RR140 pKa = 11.84RR141 pKa = 11.84STAPPFVLAAAPPQHH156 pKa = 6.15RR157 pKa = 11.84TTVRR161 pKa = 11.84PRR163 pKa = 11.84RR164 pKa = 11.84STAAAPHH171 pKa = 6.33HH172 pKa = 6.66RR173 pKa = 11.84SSSPQHH179 pKa = 6.22RR180 pKa = 11.84RR181 pKa = 11.84STAPPFVLAAAPPQHH196 pKa = 6.15RR197 pKa = 11.84TTVRR201 pKa = 11.84PRR203 pKa = 11.84RR204 pKa = 11.84STAAAPHH211 pKa = 6.33HH212 pKa = 6.66RR213 pKa = 11.84SSSPQHH219 pKa = 6.22RR220 pKa = 11.84RR221 pKa = 11.84STAPPFVLAAAPPQHH236 pKa = 6.15RR237 pKa = 11.84TTVRR241 pKa = 11.84PRR243 pKa = 11.84RR244 pKa = 11.84STAAAPHH251 pKa = 6.33HH252 pKa = 6.66RR253 pKa = 11.84SSSPQHH259 pKa = 6.22RR260 pKa = 11.84RR261 pKa = 11.84STAPPFVLAAAPPQHH276 pKa = 6.15RR277 pKa = 11.84TTVRR281 pKa = 11.84PRR283 pKa = 11.84RR284 pKa = 11.84STAAAPHH291 pKa = 6.33HH292 pKa = 6.66RR293 pKa = 11.84SSSPQHH299 pKa = 6.22RR300 pKa = 11.84RR301 pKa = 11.84STAPPFVLAAAPPQYY316 pKa = 9.25RR317 pKa = 11.84TTVRR321 pKa = 11.84PRR323 pKa = 11.84RR324 pKa = 11.84STAAAPHH331 pKa = 6.07HH332 pKa = 6.49RR333 pKa = 11.84SPFGFAGIWGKK344 pKa = 10.67EE345 pKa = 3.54IGFTDD350 pKa = 3.79LGIWII355 pKa = 4.82
Molecular weight: 38.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13452360 |
40 |
5832 |
447.6 |
49.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.438 ± 0.012 | 1.951 ± 0.007 |
5.186 ± 0.01 | 6.349 ± 0.015 |
4.299 ± 0.009 | 6.666 ± 0.025 |
2.439 ± 0.007 | 5.439 ± 0.01 |
5.872 ± 0.014 | 10.019 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.483 ± 0.006 | 4.589 ± 0.011 |
4.894 ± 0.014 | 3.636 ± 0.009 |
5.145 ± 0.01 | 8.98 ± 0.018 |
4.95 ± 0.009 | 6.61 ± 0.01 |
1.314 ± 0.005 | 2.742 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
