
Ourmia melon virus (isolate Melon/Iran/VE9) (OuMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Miaviricetes; Ourlivirales; Botourmiaviridae; Ourmiavirus; Ourmia melon virus
Average proteome isoelectric point is 8.32
Get precalculated fractions of proteins
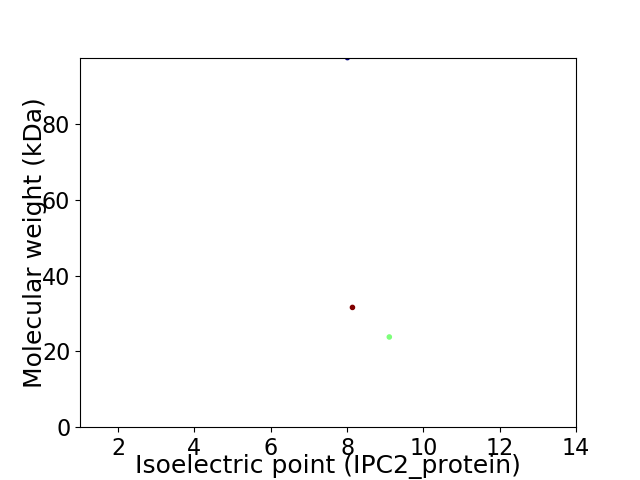
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|B3VML2|MVP_OUMVV Putative movement protein OS=Ourmia melon virus (isolate Melon/Iran/VE9) OX=652838 PE=2 SV=1
MM1 pKa = 7.82EE2 pKa = 5.55PRR4 pKa = 11.84QEE6 pKa = 4.18LLDD9 pKa = 3.7PEE11 pKa = 4.51RR12 pKa = 11.84VRR14 pKa = 11.84EE15 pKa = 3.8EE16 pKa = 3.53AKK18 pKa = 10.68RR19 pKa = 11.84IVRR22 pKa = 11.84WLCGLVDD29 pKa = 3.87RR30 pKa = 11.84TGKK33 pKa = 10.53LPAGDD38 pKa = 3.86YY39 pKa = 9.82QGKK42 pKa = 9.24ILNTVSEE49 pKa = 3.96ICKK52 pKa = 10.04RR53 pKa = 11.84SKK55 pKa = 10.75LPCGEE60 pKa = 4.03LAEE63 pKa = 4.7ALTKK67 pKa = 10.76GRR69 pKa = 11.84LTRR72 pKa = 11.84SLVDD76 pKa = 3.39YY77 pKa = 11.37SEE79 pKa = 5.24LLISNLVVGYY89 pKa = 10.15FDD91 pKa = 3.61VLEE94 pKa = 4.69IYY96 pKa = 10.31LGKK99 pKa = 10.46QSVRR103 pKa = 11.84LSDD106 pKa = 3.45IRR108 pKa = 11.84EE109 pKa = 4.05MACKK113 pKa = 9.06YY114 pKa = 8.81TFYY117 pKa = 10.79AINRR121 pKa = 11.84RR122 pKa = 11.84LEE124 pKa = 4.3DD125 pKa = 3.67YY126 pKa = 11.03IKK128 pKa = 10.78FQTAWLQARR137 pKa = 11.84AMRR140 pKa = 11.84DD141 pKa = 3.31VTPEE145 pKa = 3.77PEE147 pKa = 4.2APSWLNEE154 pKa = 3.6GFGRR158 pKa = 11.84CFSALLNRR166 pKa = 11.84KK167 pKa = 7.83VHH169 pKa = 5.78LRR171 pKa = 11.84CVLRR175 pKa = 11.84ARR177 pKa = 11.84PNDD180 pKa = 3.3VSLAASLYY188 pKa = 7.41QVKK191 pKa = 9.61RR192 pKa = 11.84VAPPLPDD199 pKa = 3.67DD200 pKa = 4.12QIEE203 pKa = 4.33KK204 pKa = 10.58NLEE207 pKa = 3.79KK208 pKa = 11.01SLDD211 pKa = 3.62RR212 pKa = 11.84LTKK215 pKa = 10.31DD216 pKa = 3.47EE217 pKa = 4.59EE218 pKa = 4.26PAGVDD223 pKa = 3.53EE224 pKa = 5.91PFLEE228 pKa = 4.44DD229 pKa = 5.08LKK231 pKa = 11.02RR232 pKa = 11.84EE233 pKa = 4.37CKK235 pKa = 9.35RR236 pKa = 11.84TVDD239 pKa = 3.8EE240 pKa = 4.41LVQNARR246 pKa = 11.84RR247 pKa = 11.84EE248 pKa = 4.0GWNRR252 pKa = 11.84KK253 pKa = 7.82ISRR256 pKa = 11.84DD257 pKa = 3.73CFPSQSAAFEE267 pKa = 4.21NPISKK272 pKa = 10.31GGQLGQLVKK281 pKa = 10.9EE282 pKa = 4.43NNTPRR287 pKa = 11.84LPVLLGMFEE296 pKa = 4.18YY297 pKa = 10.19KK298 pKa = 10.76GRR300 pKa = 11.84VTPVYY305 pKa = 10.46GWADD309 pKa = 3.78DD310 pKa = 3.84GDD312 pKa = 4.49TILSDD317 pKa = 3.64EE318 pKa = 4.24EE319 pKa = 4.22LGRR322 pKa = 11.84EE323 pKa = 4.01VPAALKK329 pKa = 10.31CRR331 pKa = 11.84RR332 pKa = 11.84SPVLEE337 pKa = 4.0PFKK340 pKa = 11.18VRR342 pKa = 11.84VITMGPAVQYY352 pKa = 9.61YY353 pKa = 8.97RR354 pKa = 11.84ARR356 pKa = 11.84RR357 pKa = 11.84VQGCLWDD364 pKa = 4.06LLKK367 pKa = 9.76HH368 pKa = 5.33TRR370 pKa = 11.84CTHH373 pKa = 5.7LPNRR377 pKa = 11.84PVEE380 pKa = 3.97EE381 pKa = 4.15SDD383 pKa = 2.7IGFYY387 pKa = 9.64VRR389 pKa = 11.84RR390 pKa = 11.84RR391 pKa = 11.84GADD394 pKa = 3.14LFRR397 pKa = 11.84GEE399 pKa = 4.25EE400 pKa = 4.09VPYY403 pKa = 11.08VSGDD407 pKa = 3.4YY408 pKa = 10.76SAATDD413 pKa = 3.91NLHH416 pKa = 7.25PDD418 pKa = 3.57LSLSVVDD425 pKa = 4.89RR426 pKa = 11.84VCDD429 pKa = 3.86HH430 pKa = 6.93LLSDD434 pKa = 5.28DD435 pKa = 4.35NRR437 pKa = 11.84PLDD440 pKa = 3.99PVSPWRR446 pKa = 11.84VLFHH450 pKa = 6.57RR451 pKa = 11.84VLVGHH456 pKa = 7.54RR457 pKa = 11.84IYY459 pKa = 11.08DD460 pKa = 3.89GNSSRR465 pKa = 11.84NTEE468 pKa = 3.65VAAQSWGQLMGSPLSFPVLCIVNLAVTRR496 pKa = 11.84YY497 pKa = 9.37VLEE500 pKa = 4.32KK501 pKa = 9.58ACGRR505 pKa = 11.84IVTLEE510 pKa = 3.71EE511 pKa = 3.98SGILVNGDD519 pKa = 4.36DD520 pKa = 3.68ILFRR524 pKa = 11.84CPEE527 pKa = 3.81RR528 pKa = 11.84TIPFWTRR535 pKa = 11.84MVTIAGLSPSPGKK548 pKa = 10.43NFVSYY553 pKa = 9.85RR554 pKa = 11.84YY555 pKa = 9.28CQLNSEE561 pKa = 4.9LYY563 pKa = 10.88DD564 pKa = 3.49MSGSRR569 pKa = 11.84AEE571 pKa = 3.9YY572 pKa = 10.78LPFIKK577 pKa = 10.62ANLIYY582 pKa = 9.25GTLARR587 pKa = 11.84GCEE590 pKa = 4.04RR591 pKa = 11.84KK592 pKa = 9.52RR593 pKa = 11.84AADD596 pKa = 3.62LCYY599 pKa = 10.9GDD601 pKa = 3.79TTTEE605 pKa = 3.99GGTFGHH611 pKa = 7.55RR612 pKa = 11.84ARR614 pKa = 11.84ALIKK618 pKa = 10.86GFGPDD623 pKa = 3.11MQDD626 pKa = 2.89RR627 pKa = 11.84LMSRR631 pKa = 11.84FLHH634 pKa = 6.08SIKK637 pKa = 10.69GFLEE641 pKa = 4.55KK642 pKa = 10.05IPEE645 pKa = 4.27VSWFIHH651 pKa = 5.46PRR653 pKa = 11.84YY654 pKa = 9.95GGLGLPLTRR663 pKa = 11.84PVTHH667 pKa = 6.48NPYY670 pKa = 9.89HH671 pKa = 6.24LRR673 pKa = 11.84IAAYY677 pKa = 9.05LSCGGEE683 pKa = 3.89QSQEE687 pKa = 3.97ARR689 pKa = 11.84CMMQWLSAPTKK700 pKa = 10.12SFNAATLLRR709 pKa = 11.84ILEE712 pKa = 4.1VARR715 pKa = 11.84NCKK718 pKa = 9.93VPFRR722 pKa = 11.84KK723 pKa = 9.99VPFALLHH730 pKa = 5.59RR731 pKa = 11.84AEE733 pKa = 4.28AAGVDD738 pKa = 3.98LEE740 pKa = 4.22ALFRR744 pKa = 11.84KK745 pKa = 9.81ALLRR749 pKa = 11.84SAPRR753 pKa = 11.84LGVEE757 pKa = 4.25YY758 pKa = 9.68PSNEE762 pKa = 4.07SGDD765 pKa = 3.82MQRR768 pKa = 11.84LGDD771 pKa = 3.47WRR773 pKa = 11.84RR774 pKa = 11.84FFRR777 pKa = 11.84DD778 pKa = 3.06VGRR781 pKa = 11.84KK782 pKa = 8.27AARR785 pKa = 11.84TRR787 pKa = 11.84CRR789 pKa = 11.84SGTADD794 pKa = 3.2EE795 pKa = 4.51RR796 pKa = 11.84KK797 pKa = 10.28GLFLMSPDD805 pKa = 3.64NAVKK809 pKa = 10.43GPQYY813 pKa = 10.98EE814 pKa = 4.97YY815 pKa = 10.4IFDD818 pKa = 3.98WASHH822 pKa = 4.79NMGGNIWDD830 pKa = 3.72PSYY833 pKa = 11.38KK834 pKa = 10.02FRR836 pKa = 11.84ADD838 pKa = 3.36PFSSSEE844 pKa = 3.92SDD846 pKa = 3.09EE847 pKa = 4.18PRR849 pKa = 11.84AKK851 pKa = 10.49EE852 pKa = 3.52IGPNRR857 pKa = 11.84GPEE860 pKa = 3.7
MM1 pKa = 7.82EE2 pKa = 5.55PRR4 pKa = 11.84QEE6 pKa = 4.18LLDD9 pKa = 3.7PEE11 pKa = 4.51RR12 pKa = 11.84VRR14 pKa = 11.84EE15 pKa = 3.8EE16 pKa = 3.53AKK18 pKa = 10.68RR19 pKa = 11.84IVRR22 pKa = 11.84WLCGLVDD29 pKa = 3.87RR30 pKa = 11.84TGKK33 pKa = 10.53LPAGDD38 pKa = 3.86YY39 pKa = 9.82QGKK42 pKa = 9.24ILNTVSEE49 pKa = 3.96ICKK52 pKa = 10.04RR53 pKa = 11.84SKK55 pKa = 10.75LPCGEE60 pKa = 4.03LAEE63 pKa = 4.7ALTKK67 pKa = 10.76GRR69 pKa = 11.84LTRR72 pKa = 11.84SLVDD76 pKa = 3.39YY77 pKa = 11.37SEE79 pKa = 5.24LLISNLVVGYY89 pKa = 10.15FDD91 pKa = 3.61VLEE94 pKa = 4.69IYY96 pKa = 10.31LGKK99 pKa = 10.46QSVRR103 pKa = 11.84LSDD106 pKa = 3.45IRR108 pKa = 11.84EE109 pKa = 4.05MACKK113 pKa = 9.06YY114 pKa = 8.81TFYY117 pKa = 10.79AINRR121 pKa = 11.84RR122 pKa = 11.84LEE124 pKa = 4.3DD125 pKa = 3.67YY126 pKa = 11.03IKK128 pKa = 10.78FQTAWLQARR137 pKa = 11.84AMRR140 pKa = 11.84DD141 pKa = 3.31VTPEE145 pKa = 3.77PEE147 pKa = 4.2APSWLNEE154 pKa = 3.6GFGRR158 pKa = 11.84CFSALLNRR166 pKa = 11.84KK167 pKa = 7.83VHH169 pKa = 5.78LRR171 pKa = 11.84CVLRR175 pKa = 11.84ARR177 pKa = 11.84PNDD180 pKa = 3.3VSLAASLYY188 pKa = 7.41QVKK191 pKa = 9.61RR192 pKa = 11.84VAPPLPDD199 pKa = 3.67DD200 pKa = 4.12QIEE203 pKa = 4.33KK204 pKa = 10.58NLEE207 pKa = 3.79KK208 pKa = 11.01SLDD211 pKa = 3.62RR212 pKa = 11.84LTKK215 pKa = 10.31DD216 pKa = 3.47EE217 pKa = 4.59EE218 pKa = 4.26PAGVDD223 pKa = 3.53EE224 pKa = 5.91PFLEE228 pKa = 4.44DD229 pKa = 5.08LKK231 pKa = 11.02RR232 pKa = 11.84EE233 pKa = 4.37CKK235 pKa = 9.35RR236 pKa = 11.84TVDD239 pKa = 3.8EE240 pKa = 4.41LVQNARR246 pKa = 11.84RR247 pKa = 11.84EE248 pKa = 4.0GWNRR252 pKa = 11.84KK253 pKa = 7.82ISRR256 pKa = 11.84DD257 pKa = 3.73CFPSQSAAFEE267 pKa = 4.21NPISKK272 pKa = 10.31GGQLGQLVKK281 pKa = 10.9EE282 pKa = 4.43NNTPRR287 pKa = 11.84LPVLLGMFEE296 pKa = 4.18YY297 pKa = 10.19KK298 pKa = 10.76GRR300 pKa = 11.84VTPVYY305 pKa = 10.46GWADD309 pKa = 3.78DD310 pKa = 3.84GDD312 pKa = 4.49TILSDD317 pKa = 3.64EE318 pKa = 4.24EE319 pKa = 4.22LGRR322 pKa = 11.84EE323 pKa = 4.01VPAALKK329 pKa = 10.31CRR331 pKa = 11.84RR332 pKa = 11.84SPVLEE337 pKa = 4.0PFKK340 pKa = 11.18VRR342 pKa = 11.84VITMGPAVQYY352 pKa = 9.61YY353 pKa = 8.97RR354 pKa = 11.84ARR356 pKa = 11.84RR357 pKa = 11.84VQGCLWDD364 pKa = 4.06LLKK367 pKa = 9.76HH368 pKa = 5.33TRR370 pKa = 11.84CTHH373 pKa = 5.7LPNRR377 pKa = 11.84PVEE380 pKa = 3.97EE381 pKa = 4.15SDD383 pKa = 2.7IGFYY387 pKa = 9.64VRR389 pKa = 11.84RR390 pKa = 11.84RR391 pKa = 11.84GADD394 pKa = 3.14LFRR397 pKa = 11.84GEE399 pKa = 4.25EE400 pKa = 4.09VPYY403 pKa = 11.08VSGDD407 pKa = 3.4YY408 pKa = 10.76SAATDD413 pKa = 3.91NLHH416 pKa = 7.25PDD418 pKa = 3.57LSLSVVDD425 pKa = 4.89RR426 pKa = 11.84VCDD429 pKa = 3.86HH430 pKa = 6.93LLSDD434 pKa = 5.28DD435 pKa = 4.35NRR437 pKa = 11.84PLDD440 pKa = 3.99PVSPWRR446 pKa = 11.84VLFHH450 pKa = 6.57RR451 pKa = 11.84VLVGHH456 pKa = 7.54RR457 pKa = 11.84IYY459 pKa = 11.08DD460 pKa = 3.89GNSSRR465 pKa = 11.84NTEE468 pKa = 3.65VAAQSWGQLMGSPLSFPVLCIVNLAVTRR496 pKa = 11.84YY497 pKa = 9.37VLEE500 pKa = 4.32KK501 pKa = 9.58ACGRR505 pKa = 11.84IVTLEE510 pKa = 3.71EE511 pKa = 3.98SGILVNGDD519 pKa = 4.36DD520 pKa = 3.68ILFRR524 pKa = 11.84CPEE527 pKa = 3.81RR528 pKa = 11.84TIPFWTRR535 pKa = 11.84MVTIAGLSPSPGKK548 pKa = 10.43NFVSYY553 pKa = 9.85RR554 pKa = 11.84YY555 pKa = 9.28CQLNSEE561 pKa = 4.9LYY563 pKa = 10.88DD564 pKa = 3.49MSGSRR569 pKa = 11.84AEE571 pKa = 3.9YY572 pKa = 10.78LPFIKK577 pKa = 10.62ANLIYY582 pKa = 9.25GTLARR587 pKa = 11.84GCEE590 pKa = 4.04RR591 pKa = 11.84KK592 pKa = 9.52RR593 pKa = 11.84AADD596 pKa = 3.62LCYY599 pKa = 10.9GDD601 pKa = 3.79TTTEE605 pKa = 3.99GGTFGHH611 pKa = 7.55RR612 pKa = 11.84ARR614 pKa = 11.84ALIKK618 pKa = 10.86GFGPDD623 pKa = 3.11MQDD626 pKa = 2.89RR627 pKa = 11.84LMSRR631 pKa = 11.84FLHH634 pKa = 6.08SIKK637 pKa = 10.69GFLEE641 pKa = 4.55KK642 pKa = 10.05IPEE645 pKa = 4.27VSWFIHH651 pKa = 5.46PRR653 pKa = 11.84YY654 pKa = 9.95GGLGLPLTRR663 pKa = 11.84PVTHH667 pKa = 6.48NPYY670 pKa = 9.89HH671 pKa = 6.24LRR673 pKa = 11.84IAAYY677 pKa = 9.05LSCGGEE683 pKa = 3.89QSQEE687 pKa = 3.97ARR689 pKa = 11.84CMMQWLSAPTKK700 pKa = 10.12SFNAATLLRR709 pKa = 11.84ILEE712 pKa = 4.1VARR715 pKa = 11.84NCKK718 pKa = 9.93VPFRR722 pKa = 11.84KK723 pKa = 9.99VPFALLHH730 pKa = 5.59RR731 pKa = 11.84AEE733 pKa = 4.28AAGVDD738 pKa = 3.98LEE740 pKa = 4.22ALFRR744 pKa = 11.84KK745 pKa = 9.81ALLRR749 pKa = 11.84SAPRR753 pKa = 11.84LGVEE757 pKa = 4.25YY758 pKa = 9.68PSNEE762 pKa = 4.07SGDD765 pKa = 3.82MQRR768 pKa = 11.84LGDD771 pKa = 3.47WRR773 pKa = 11.84RR774 pKa = 11.84FFRR777 pKa = 11.84DD778 pKa = 3.06VGRR781 pKa = 11.84KK782 pKa = 8.27AARR785 pKa = 11.84TRR787 pKa = 11.84CRR789 pKa = 11.84SGTADD794 pKa = 3.2EE795 pKa = 4.51RR796 pKa = 11.84KK797 pKa = 10.28GLFLMSPDD805 pKa = 3.64NAVKK809 pKa = 10.43GPQYY813 pKa = 10.98EE814 pKa = 4.97YY815 pKa = 10.4IFDD818 pKa = 3.98WASHH822 pKa = 4.79NMGGNIWDD830 pKa = 3.72PSYY833 pKa = 11.38KK834 pKa = 10.02FRR836 pKa = 11.84ADD838 pKa = 3.36PFSSSEE844 pKa = 3.92SDD846 pKa = 3.09EE847 pKa = 4.18PRR849 pKa = 11.84AKK851 pKa = 10.49EE852 pKa = 3.52IGPNRR857 pKa = 11.84GPEE860 pKa = 3.7
Molecular weight: 97.55 kDa
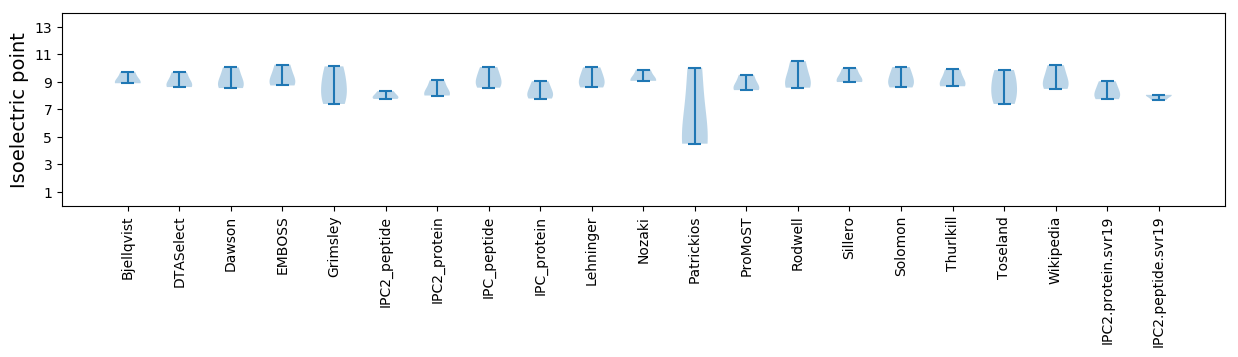
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|B3VML3|CAPSD_OUMVV Capsid protein OS=Ourmia melon virus (isolate Melon/Iran/VE9) OX=652838 PE=2 SV=1
MM1 pKa = 7.55ARR3 pKa = 11.84LPKK6 pKa = 10.25RR7 pKa = 11.84KK8 pKa = 9.26NRR10 pKa = 11.84RR11 pKa = 11.84NEE13 pKa = 3.75KK14 pKa = 10.12KK15 pKa = 10.5KK16 pKa = 10.1NANASRR22 pKa = 11.84VQNVPRR28 pKa = 11.84TFGLWKK34 pKa = 7.82STEE37 pKa = 4.11RR38 pKa = 11.84IKK40 pKa = 10.73YY41 pKa = 4.59TTEE44 pKa = 3.44LKK46 pKa = 10.75YY47 pKa = 11.19LNSKK51 pKa = 9.58CRR53 pKa = 11.84AIRR56 pKa = 11.84LHH58 pKa = 7.11PDD60 pKa = 3.55LVANNSFPTYY70 pKa = 10.0CSAWKK75 pKa = 9.49IDD77 pKa = 3.42QVEE80 pKa = 4.4FEE82 pKa = 4.56FVSYY86 pKa = 9.52MSPLAGHH93 pKa = 6.13VGCVFFVVIPAKK105 pKa = 10.37GLNSRR110 pKa = 11.84ISADD114 pKa = 3.46EE115 pKa = 4.39AEE117 pKa = 4.67SLQSAILWDD126 pKa = 3.52EE127 pKa = 4.0KK128 pKa = 11.12GRR130 pKa = 11.84LKK132 pKa = 9.41ITPISGPISRR142 pKa = 11.84HH143 pKa = 4.37PWTNLSQVVTPPQIPKK159 pKa = 10.5GSTDD163 pKa = 3.66GEE165 pKa = 4.24RR166 pKa = 11.84QDD168 pKa = 4.35LQSGYY173 pKa = 10.67YY174 pKa = 10.53LIFDD178 pKa = 3.63SRR180 pKa = 11.84KK181 pKa = 10.0LFGKK185 pKa = 10.27DD186 pKa = 3.26LVDD189 pKa = 3.54KK190 pKa = 11.1QSVLGEE196 pKa = 4.05LSLTITATYY205 pKa = 6.88WTSLSS210 pKa = 3.57
MM1 pKa = 7.55ARR3 pKa = 11.84LPKK6 pKa = 10.25RR7 pKa = 11.84KK8 pKa = 9.26NRR10 pKa = 11.84RR11 pKa = 11.84NEE13 pKa = 3.75KK14 pKa = 10.12KK15 pKa = 10.5KK16 pKa = 10.1NANASRR22 pKa = 11.84VQNVPRR28 pKa = 11.84TFGLWKK34 pKa = 7.82STEE37 pKa = 4.11RR38 pKa = 11.84IKK40 pKa = 10.73YY41 pKa = 4.59TTEE44 pKa = 3.44LKK46 pKa = 10.75YY47 pKa = 11.19LNSKK51 pKa = 9.58CRR53 pKa = 11.84AIRR56 pKa = 11.84LHH58 pKa = 7.11PDD60 pKa = 3.55LVANNSFPTYY70 pKa = 10.0CSAWKK75 pKa = 9.49IDD77 pKa = 3.42QVEE80 pKa = 4.4FEE82 pKa = 4.56FVSYY86 pKa = 9.52MSPLAGHH93 pKa = 6.13VGCVFFVVIPAKK105 pKa = 10.37GLNSRR110 pKa = 11.84ISADD114 pKa = 3.46EE115 pKa = 4.39AEE117 pKa = 4.67SLQSAILWDD126 pKa = 3.52EE127 pKa = 4.0KK128 pKa = 11.12GRR130 pKa = 11.84LKK132 pKa = 9.41ITPISGPISRR142 pKa = 11.84HH143 pKa = 4.37PWTNLSQVVTPPQIPKK159 pKa = 10.5GSTDD163 pKa = 3.66GEE165 pKa = 4.24RR166 pKa = 11.84QDD168 pKa = 4.35LQSGYY173 pKa = 10.67YY174 pKa = 10.53LIFDD178 pKa = 3.63SRR180 pKa = 11.84KK181 pKa = 10.0LFGKK185 pKa = 10.27DD186 pKa = 3.26LVDD189 pKa = 3.54KK190 pKa = 11.1QSVLGEE196 pKa = 4.05LSLTITATYY205 pKa = 6.88WTSLSS210 pKa = 3.57
Molecular weight: 23.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1358 |
210 |
860 |
452.7 |
50.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.627 ± 0.315 | 2.062 ± 0.457 |
5.523 ± 0.323 | 6.112 ± 0.667 |
3.24 ± 0.772 | 6.922 ± 0.42 |
1.841 ± 0.325 | 4.86 ± 1.115 |
5.081 ± 0.753 | 10.309 ± 0.455 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.694 ± 0.201 | 3.387 ± 0.498 |
6.701 ± 0.688 | 3.166 ± 0.573 |
8.91 ± 0.966 | 7.585 ± 1.055 |
4.934 ± 1.024 | 6.186 ± 0.486 |
1.62 ± 0.24 | 3.24 ± 0.29 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
