
Giant panda circovirus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; unclassified Circovirus
Average proteome isoelectric point is 7.55
Get precalculated fractions of proteins
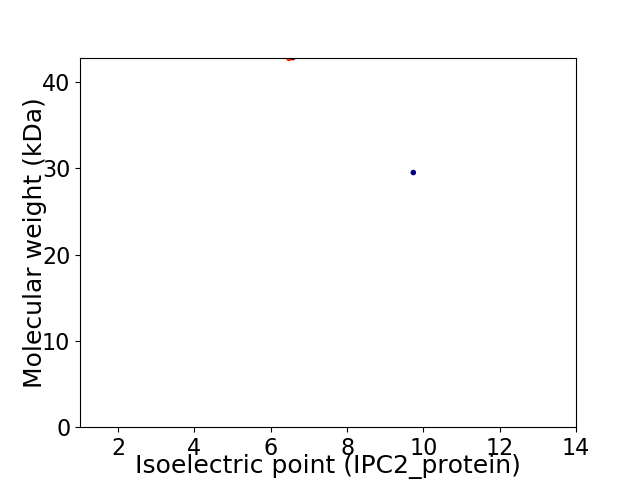
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
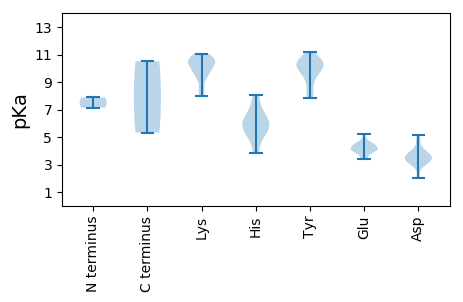
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A220IGT3|A0A220IGT3_9CIRC Capsid protein OS=Giant panda circovirus 4 OX=2016459 PE=4 SV=1
MM1 pKa = 7.14ITPLILQINTMSRR14 pKa = 11.84LRR16 pKa = 11.84NCCFTINNIDD26 pKa = 3.58ANNNIEE32 pKa = 4.22NYY34 pKa = 10.25FNFPIDD40 pKa = 3.49GVKK43 pKa = 9.87YY44 pKa = 10.5CIYY47 pKa = 10.37QKK49 pKa = 11.05EE50 pKa = 4.57KK51 pKa = 9.26VTRR54 pKa = 11.84DD55 pKa = 3.14HH56 pKa = 6.06LQGYY60 pKa = 9.6IEE62 pKa = 4.59FEE64 pKa = 4.17APKK67 pKa = 10.35SQAQIKK73 pKa = 10.09DD74 pKa = 3.38ILGKK78 pKa = 9.57EE79 pKa = 3.58AHH81 pKa = 5.86IEE83 pKa = 4.01KK84 pKa = 10.49RR85 pKa = 11.84RR86 pKa = 11.84GTALQAQQYY95 pKa = 9.58CMKK98 pKa = 10.19NDD100 pKa = 3.33SRR102 pKa = 11.84VSGPYY107 pKa = 8.2EE108 pKa = 3.81FGVISHH114 pKa = 5.72QGKK117 pKa = 10.18RR118 pKa = 11.84SDD120 pKa = 3.76LKK122 pKa = 10.95EE123 pKa = 3.51IADD126 pKa = 3.97RR127 pKa = 11.84FRR129 pKa = 11.84SGEE132 pKa = 4.21SIKK135 pKa = 10.79DD136 pKa = 3.33IIDD139 pKa = 3.69GNEE142 pKa = 4.01TQFVRR147 pKa = 11.84YY148 pKa = 9.69GRR150 pKa = 11.84GLRR153 pKa = 11.84EE154 pKa = 4.14LQALLQPRR162 pKa = 11.84PRR164 pKa = 11.84SYY166 pKa = 9.82TKK168 pKa = 10.71KK169 pKa = 9.85EE170 pKa = 4.0VVVLWGPPGVGKK182 pKa = 10.6SYY184 pKa = 10.4FVNHH188 pKa = 5.84IMEE191 pKa = 4.99LRR193 pKa = 11.84QKK195 pKa = 10.65LLYY198 pKa = 10.02IKK200 pKa = 10.56SPNTKK205 pKa = 8.41WWDD208 pKa = 3.46NYY210 pKa = 10.48QGEE213 pKa = 4.55SDD215 pKa = 3.52ILLDD219 pKa = 4.0EE220 pKa = 5.2FPGTMTARR228 pKa = 11.84DD229 pKa = 3.47AKK231 pKa = 10.61IILGEE236 pKa = 4.15VNGPLEE242 pKa = 4.34TKK244 pKa = 10.39GGSFTTDD251 pKa = 2.04KK252 pKa = 10.86MEE254 pKa = 4.55RR255 pKa = 11.84FWITSNYY262 pKa = 8.98EE263 pKa = 3.38PGMWFEE269 pKa = 4.0NAKK272 pKa = 10.82DD273 pKa = 3.43IDD275 pKa = 3.7RR276 pKa = 11.84SAVQRR281 pKa = 11.84EE282 pKa = 4.33VNGPLEE288 pKa = 4.35TKK290 pKa = 10.39GGSFTTDD297 pKa = 2.04KK298 pKa = 10.86MEE300 pKa = 4.55RR301 pKa = 11.84FWITSNYY308 pKa = 8.98EE309 pKa = 3.38PGMWFEE315 pKa = 4.0NAKK318 pKa = 10.82DD319 pKa = 3.43IDD321 pKa = 3.7RR322 pKa = 11.84SAVQRR327 pKa = 11.84RR328 pKa = 11.84LTTICHH334 pKa = 3.83VTEE337 pKa = 4.19WQDD340 pKa = 3.22TAEE343 pKa = 3.98IFYY346 pKa = 10.81KK347 pKa = 10.2FFPPPSIQFGPPYY360 pKa = 10.6DD361 pKa = 3.8EE362 pKa = 4.11TQNIYY367 pKa = 10.43KK368 pKa = 10.51
MM1 pKa = 7.14ITPLILQINTMSRR14 pKa = 11.84LRR16 pKa = 11.84NCCFTINNIDD26 pKa = 3.58ANNNIEE32 pKa = 4.22NYY34 pKa = 10.25FNFPIDD40 pKa = 3.49GVKK43 pKa = 9.87YY44 pKa = 10.5CIYY47 pKa = 10.37QKK49 pKa = 11.05EE50 pKa = 4.57KK51 pKa = 9.26VTRR54 pKa = 11.84DD55 pKa = 3.14HH56 pKa = 6.06LQGYY60 pKa = 9.6IEE62 pKa = 4.59FEE64 pKa = 4.17APKK67 pKa = 10.35SQAQIKK73 pKa = 10.09DD74 pKa = 3.38ILGKK78 pKa = 9.57EE79 pKa = 3.58AHH81 pKa = 5.86IEE83 pKa = 4.01KK84 pKa = 10.49RR85 pKa = 11.84RR86 pKa = 11.84GTALQAQQYY95 pKa = 9.58CMKK98 pKa = 10.19NDD100 pKa = 3.33SRR102 pKa = 11.84VSGPYY107 pKa = 8.2EE108 pKa = 3.81FGVISHH114 pKa = 5.72QGKK117 pKa = 10.18RR118 pKa = 11.84SDD120 pKa = 3.76LKK122 pKa = 10.95EE123 pKa = 3.51IADD126 pKa = 3.97RR127 pKa = 11.84FRR129 pKa = 11.84SGEE132 pKa = 4.21SIKK135 pKa = 10.79DD136 pKa = 3.33IIDD139 pKa = 3.69GNEE142 pKa = 4.01TQFVRR147 pKa = 11.84YY148 pKa = 9.69GRR150 pKa = 11.84GLRR153 pKa = 11.84EE154 pKa = 4.14LQALLQPRR162 pKa = 11.84PRR164 pKa = 11.84SYY166 pKa = 9.82TKK168 pKa = 10.71KK169 pKa = 9.85EE170 pKa = 4.0VVVLWGPPGVGKK182 pKa = 10.6SYY184 pKa = 10.4FVNHH188 pKa = 5.84IMEE191 pKa = 4.99LRR193 pKa = 11.84QKK195 pKa = 10.65LLYY198 pKa = 10.02IKK200 pKa = 10.56SPNTKK205 pKa = 8.41WWDD208 pKa = 3.46NYY210 pKa = 10.48QGEE213 pKa = 4.55SDD215 pKa = 3.52ILLDD219 pKa = 4.0EE220 pKa = 5.2FPGTMTARR228 pKa = 11.84DD229 pKa = 3.47AKK231 pKa = 10.61IILGEE236 pKa = 4.15VNGPLEE242 pKa = 4.34TKK244 pKa = 10.39GGSFTTDD251 pKa = 2.04KK252 pKa = 10.86MEE254 pKa = 4.55RR255 pKa = 11.84FWITSNYY262 pKa = 8.98EE263 pKa = 3.38PGMWFEE269 pKa = 4.0NAKK272 pKa = 10.82DD273 pKa = 3.43IDD275 pKa = 3.7RR276 pKa = 11.84SAVQRR281 pKa = 11.84EE282 pKa = 4.33VNGPLEE288 pKa = 4.35TKK290 pKa = 10.39GGSFTTDD297 pKa = 2.04KK298 pKa = 10.86MEE300 pKa = 4.55RR301 pKa = 11.84FWITSNYY308 pKa = 8.98EE309 pKa = 3.38PGMWFEE315 pKa = 4.0NAKK318 pKa = 10.82DD319 pKa = 3.43IDD321 pKa = 3.7RR322 pKa = 11.84SAVQRR327 pKa = 11.84RR328 pKa = 11.84LTTICHH334 pKa = 3.83VTEE337 pKa = 4.19WQDD340 pKa = 3.22TAEE343 pKa = 3.98IFYY346 pKa = 10.81KK347 pKa = 10.2FFPPPSIQFGPPYY360 pKa = 10.6DD361 pKa = 3.8EE362 pKa = 4.11TQNIYY367 pKa = 10.43KK368 pKa = 10.51
Molecular weight: 42.72 kDa
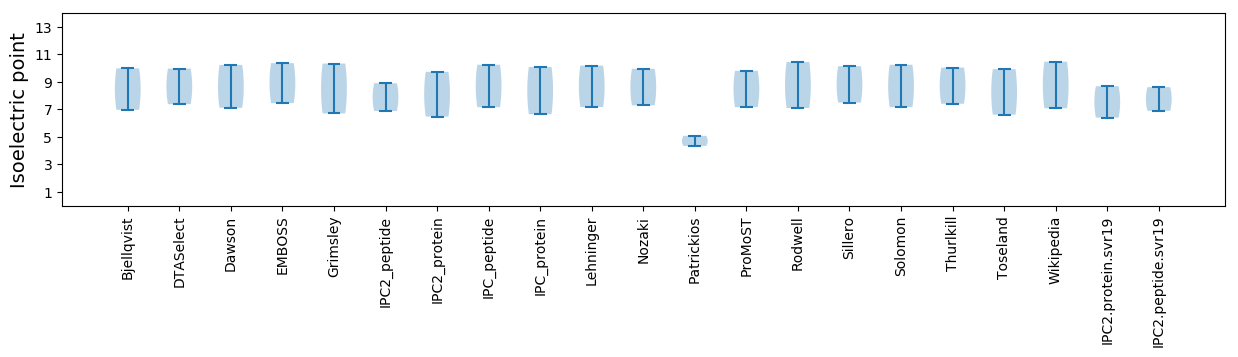
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A220IGT3|A0A220IGT3_9CIRC Capsid protein OS=Giant panda circovirus 4 OX=2016459 PE=4 SV=1
MM1 pKa = 7.87RR2 pKa = 11.84KK3 pKa = 9.42FFINFFLPLRR13 pKa = 11.84YY14 pKa = 9.9NLDD17 pKa = 3.03HH18 pKa = 8.08RR19 pKa = 11.84MMKK22 pKa = 10.27RR23 pKa = 11.84KK24 pKa = 8.16TYY26 pKa = 7.83TNRR29 pKa = 11.84SYY31 pKa = 10.2RR32 pKa = 11.84TTYY35 pKa = 10.13KK36 pKa = 9.72RR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 8.25YY40 pKa = 8.38GTNRR44 pKa = 11.84KK45 pKa = 8.6VPKK48 pKa = 9.5PLRR51 pKa = 11.84TAIKK55 pKa = 9.18KK56 pKa = 7.99TMNRR60 pKa = 11.84QLEE63 pKa = 4.48TKK65 pKa = 9.84KK66 pKa = 10.56CYY68 pKa = 8.16QQGMTVMQANFQANTMYY85 pKa = 10.51TDD87 pKa = 4.2NILYY91 pKa = 9.19NIPQNATEE99 pKa = 4.53GGRR102 pKa = 11.84IGDD105 pKa = 4.7RR106 pKa = 11.84INISKK111 pKa = 10.21VVISGEE117 pKa = 4.34FQCQPTDD124 pKa = 3.16NVFWHH129 pKa = 6.72LYY131 pKa = 8.05IFRR134 pKa = 11.84SALYY138 pKa = 10.02NDD140 pKa = 3.71NTTPPLYY147 pKa = 10.88GIMPYY152 pKa = 10.59SRR154 pKa = 11.84FYY156 pKa = 10.98HH157 pKa = 5.51PVGSIVNPNVDD168 pKa = 5.15LPDD171 pKa = 4.91PDD173 pKa = 3.11QVRR176 pKa = 11.84ILKK179 pKa = 9.48YY180 pKa = 10.21KK181 pKa = 10.38RR182 pKa = 11.84IRR184 pKa = 11.84VDD186 pKa = 3.05PANNGGICIRR196 pKa = 11.84RR197 pKa = 11.84FYY199 pKa = 11.19LSVNMKK205 pKa = 9.76NAAYY209 pKa = 9.92QYY211 pKa = 9.34ATDD214 pKa = 4.04GSSYY218 pKa = 9.73GRR220 pKa = 11.84NYY222 pKa = 10.74NLYY225 pKa = 10.11FGVICRR231 pKa = 11.84GQEE234 pKa = 3.94GNQLNITEE242 pKa = 3.89QRR244 pKa = 11.84IFYY247 pKa = 10.52KK248 pKa = 10.75DD249 pKa = 2.89AA250 pKa = 5.33
MM1 pKa = 7.87RR2 pKa = 11.84KK3 pKa = 9.42FFINFFLPLRR13 pKa = 11.84YY14 pKa = 9.9NLDD17 pKa = 3.03HH18 pKa = 8.08RR19 pKa = 11.84MMKK22 pKa = 10.27RR23 pKa = 11.84KK24 pKa = 8.16TYY26 pKa = 7.83TNRR29 pKa = 11.84SYY31 pKa = 10.2RR32 pKa = 11.84TTYY35 pKa = 10.13KK36 pKa = 9.72RR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 8.25YY40 pKa = 8.38GTNRR44 pKa = 11.84KK45 pKa = 8.6VPKK48 pKa = 9.5PLRR51 pKa = 11.84TAIKK55 pKa = 9.18KK56 pKa = 7.99TMNRR60 pKa = 11.84QLEE63 pKa = 4.48TKK65 pKa = 9.84KK66 pKa = 10.56CYY68 pKa = 8.16QQGMTVMQANFQANTMYY85 pKa = 10.51TDD87 pKa = 4.2NILYY91 pKa = 9.19NIPQNATEE99 pKa = 4.53GGRR102 pKa = 11.84IGDD105 pKa = 4.7RR106 pKa = 11.84INISKK111 pKa = 10.21VVISGEE117 pKa = 4.34FQCQPTDD124 pKa = 3.16NVFWHH129 pKa = 6.72LYY131 pKa = 8.05IFRR134 pKa = 11.84SALYY138 pKa = 10.02NDD140 pKa = 3.71NTTPPLYY147 pKa = 10.88GIMPYY152 pKa = 10.59SRR154 pKa = 11.84FYY156 pKa = 10.98HH157 pKa = 5.51PVGSIVNPNVDD168 pKa = 5.15LPDD171 pKa = 4.91PDD173 pKa = 3.11QVRR176 pKa = 11.84ILKK179 pKa = 9.48YY180 pKa = 10.21KK181 pKa = 10.38RR182 pKa = 11.84IRR184 pKa = 11.84VDD186 pKa = 3.05PANNGGICIRR196 pKa = 11.84RR197 pKa = 11.84FYY199 pKa = 11.19LSVNMKK205 pKa = 9.76NAAYY209 pKa = 9.92QYY211 pKa = 9.34ATDD214 pKa = 4.04GSSYY218 pKa = 9.73GRR220 pKa = 11.84NYY222 pKa = 10.74NLYY225 pKa = 10.11FGVICRR231 pKa = 11.84GQEE234 pKa = 3.94GNQLNITEE242 pKa = 3.89QRR244 pKa = 11.84IFYY247 pKa = 10.52KK248 pKa = 10.75DD249 pKa = 2.89AA250 pKa = 5.33
Molecular weight: 29.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
618 |
250 |
368 |
309.0 |
36.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.045 ± 0.026 | 1.456 ± 0.084 |
5.178 ± 0.455 | 5.34 ± 1.952 |
5.016 ± 0.126 | 6.634 ± 0.371 |
1.294 ± 0.055 | 7.767 ± 0.331 |
6.796 ± 0.232 | 5.663 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.913 ± 0.402 | 7.282 ± 1.355 |
5.178 ± 0.013 | 5.178 ± 0.013 |
7.443 ± 1.027 | 4.531 ± 0.544 |
6.472 ± 0.191 | 4.369 ± 0.252 |
1.456 ± 0.618 | 5.987 ± 1.411 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
