
Thiohalorhabdus denitrificans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Gammaproteobacteria incertae sedis; Thiohalorhabdus
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
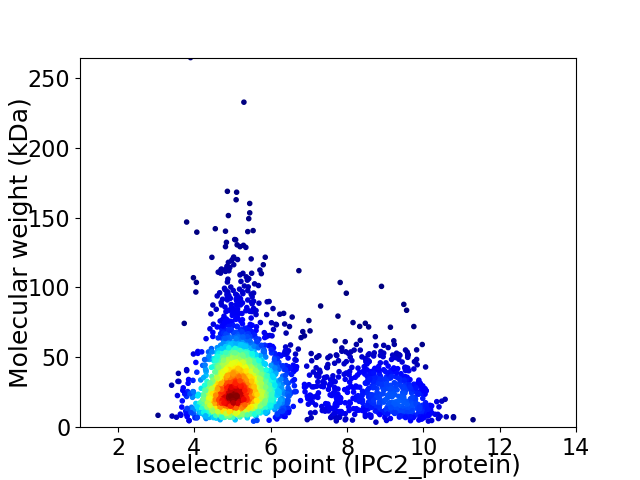
Virtual 2D-PAGE plot for 2692 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G5GVC9|A0A1G5GVC9_9GAMM Competence protein ComEA OS=Thiohalorhabdus denitrificans OX=381306 GN=SAMN05661077_2470 PE=4 SV=1
MM1 pKa = 7.67RR2 pKa = 11.84RR3 pKa = 11.84IQNGPAAEE11 pKa = 4.19TGHH14 pKa = 7.22ADD16 pKa = 3.18GTAAPGRR23 pKa = 11.84LRR25 pKa = 11.84HH26 pKa = 5.89HH27 pKa = 6.52VLPGLALGLLLLPAPAGAADD47 pKa = 4.85DD48 pKa = 4.65CPSDD52 pKa = 4.08ASTTCSVDD60 pKa = 3.06VGSSSEE66 pKa = 4.2GEE68 pKa = 3.83MGGNSDD74 pKa = 3.74TDD76 pKa = 3.37WFKK79 pKa = 10.97VTGLEE84 pKa = 3.82AGKK87 pKa = 8.57TYY89 pKa = 10.7RR90 pKa = 11.84FHH92 pKa = 7.33QKK94 pKa = 10.96GEE96 pKa = 4.23TSDD99 pKa = 3.58EE100 pKa = 4.28TLTLLDD106 pKa = 4.16PYY108 pKa = 11.4ARR110 pKa = 11.84LVDD113 pKa = 3.92ANGTVVAEE121 pKa = 4.93DD122 pKa = 4.98DD123 pKa = 4.11DD124 pKa = 4.36TGPMNNAQITYY135 pKa = 9.9SPVEE139 pKa = 4.15AEE141 pKa = 3.98LPLYY145 pKa = 10.8LEE147 pKa = 5.63ADD149 pKa = 3.97NQPDD153 pKa = 4.3DD154 pKa = 3.48PSGTYY159 pKa = 9.22RR160 pKa = 11.84VSVEE164 pKa = 3.95EE165 pKa = 5.45FVDD168 pKa = 4.03DD169 pKa = 4.16DD170 pKa = 4.42CVDD173 pKa = 3.88RR174 pKa = 11.84PHH176 pKa = 7.04TDD178 pKa = 2.84SDD180 pKa = 4.2CALEE184 pKa = 4.75LDD186 pKa = 4.98TIITWGIDD194 pKa = 3.38FAGDD198 pKa = 3.75EE199 pKa = 4.46DD200 pKa = 4.11WLKK203 pKa = 11.38ASVDD207 pKa = 3.82TSSGPHH213 pKa = 4.93YY214 pKa = 10.28QFSVANSDD222 pKa = 3.72SQDD225 pKa = 2.99VSLEE229 pKa = 3.74LRR231 pKa = 11.84AQDD234 pKa = 3.97GNLHH238 pKa = 6.22IADD241 pKa = 4.41EE242 pKa = 4.78GADD245 pKa = 3.58EE246 pKa = 4.36ATLTFTTTDD255 pKa = 3.32DD256 pKa = 3.7SNYY259 pKa = 9.01FAAVSFLSSEE269 pKa = 4.21TGDD272 pKa = 4.15FSATYY277 pKa = 8.76TNYY280 pKa = 10.79SGGCPSNPLYY290 pKa = 10.3PSALDD295 pKa = 3.47YY296 pKa = 11.65SNYY299 pKa = 8.64STDD302 pKa = 3.62YY303 pKa = 9.79QKK305 pKa = 11.17CGAEE309 pKa = 4.15APGPDD314 pKa = 3.97AGSATLSGSIEE325 pKa = 4.2PVGDD329 pKa = 3.59TDD331 pKa = 3.33WFKK334 pKa = 10.49ITGMQADD341 pKa = 3.41QSYY344 pKa = 10.42QLDD347 pKa = 4.18LTSGTDD353 pKa = 3.67GLPDD357 pKa = 4.32PLLRR361 pKa = 11.84IYY363 pKa = 10.56DD364 pKa = 3.7AEE366 pKa = 4.32GNEE369 pKa = 4.78LAVDD373 pKa = 3.83DD374 pKa = 5.39DD375 pKa = 4.67GGSKK379 pKa = 10.41EE380 pKa = 4.06DD381 pKa = 3.57AQLGFSPPADD391 pKa = 3.2GDD393 pKa = 3.99YY394 pKa = 10.49FASAEE399 pKa = 4.26SSPKK403 pKa = 9.69DD404 pKa = 3.67DD405 pKa = 3.54QRR407 pKa = 11.84GGYY410 pKa = 8.31SLEE413 pKa = 3.87VANYY417 pKa = 10.1SDD419 pKa = 4.38RR420 pKa = 11.84DD421 pKa = 3.97CAGSNATDD429 pKa = 3.65CGIKK433 pKa = 10.07IGDD436 pKa = 3.85TLVADD441 pKa = 5.75GSTSWDD447 pKa = 3.7YY448 pKa = 10.92EE449 pKa = 4.25VHH451 pKa = 6.65VSGDD455 pKa = 3.72SDD457 pKa = 3.42WFAVSGMTAGEE468 pKa = 4.27SYY470 pKa = 10.79KK471 pKa = 10.7FEE473 pKa = 4.46LRR475 pKa = 11.84NTDD478 pKa = 5.02GILQDD483 pKa = 4.61PYY485 pKa = 11.56LSVYY489 pKa = 10.43DD490 pKa = 3.99SNGEE494 pKa = 3.94EE495 pKa = 4.24LRR497 pKa = 11.84SSGLSKK503 pKa = 11.07DD504 pKa = 3.44PAQLGFTPPDD514 pKa = 3.49NGDD517 pKa = 2.94YY518 pKa = 10.37WIAAEE523 pKa = 4.9SGTEE527 pKa = 3.92GDD529 pKa = 3.13TGAYY533 pKa = 8.19EE534 pKa = 4.2LSAGKK539 pKa = 10.48FNDD542 pKa = 3.65DD543 pKa = 4.17HH544 pKa = 7.3ADD546 pKa = 3.8CATSACSPGTLPVDD560 pKa = 4.13DD561 pKa = 4.62SANGEE566 pKa = 4.27LEE568 pKa = 4.33VPGDD572 pKa = 3.84RR573 pKa = 11.84DD574 pKa = 3.18WFKK577 pKa = 11.8VDD579 pKa = 4.48LEE581 pKa = 4.36SGNYY585 pKa = 7.15YY586 pKa = 9.84TFTLSGDD593 pKa = 3.81TLGAPHH599 pKa = 6.21VRR601 pKa = 11.84LRR603 pKa = 11.84TDD605 pKa = 3.63DD606 pKa = 3.95GSILASQAASPDD618 pKa = 3.31SSEE621 pKa = 4.67AKK623 pKa = 8.86VTHH626 pKa = 6.35EE627 pKa = 4.24PGANQTVYY635 pKa = 11.11LEE637 pKa = 4.33AASVEE642 pKa = 4.25DD643 pKa = 4.64EE644 pKa = 4.34SAGTYY649 pKa = 7.63TVSATAQDD657 pKa = 3.98LSGGTGGGSSDD668 pKa = 3.5GGGSGGGGGCALAAEE683 pKa = 4.73PQPLDD688 pKa = 3.25PTLPLLAGLGLAYY701 pKa = 10.61LLRR704 pKa = 11.84RR705 pKa = 11.84GRR707 pKa = 11.84PRR709 pKa = 11.84AA710 pKa = 3.4
MM1 pKa = 7.67RR2 pKa = 11.84RR3 pKa = 11.84IQNGPAAEE11 pKa = 4.19TGHH14 pKa = 7.22ADD16 pKa = 3.18GTAAPGRR23 pKa = 11.84LRR25 pKa = 11.84HH26 pKa = 5.89HH27 pKa = 6.52VLPGLALGLLLLPAPAGAADD47 pKa = 4.85DD48 pKa = 4.65CPSDD52 pKa = 4.08ASTTCSVDD60 pKa = 3.06VGSSSEE66 pKa = 4.2GEE68 pKa = 3.83MGGNSDD74 pKa = 3.74TDD76 pKa = 3.37WFKK79 pKa = 10.97VTGLEE84 pKa = 3.82AGKK87 pKa = 8.57TYY89 pKa = 10.7RR90 pKa = 11.84FHH92 pKa = 7.33QKK94 pKa = 10.96GEE96 pKa = 4.23TSDD99 pKa = 3.58EE100 pKa = 4.28TLTLLDD106 pKa = 4.16PYY108 pKa = 11.4ARR110 pKa = 11.84LVDD113 pKa = 3.92ANGTVVAEE121 pKa = 4.93DD122 pKa = 4.98DD123 pKa = 4.11DD124 pKa = 4.36TGPMNNAQITYY135 pKa = 9.9SPVEE139 pKa = 4.15AEE141 pKa = 3.98LPLYY145 pKa = 10.8LEE147 pKa = 5.63ADD149 pKa = 3.97NQPDD153 pKa = 4.3DD154 pKa = 3.48PSGTYY159 pKa = 9.22RR160 pKa = 11.84VSVEE164 pKa = 3.95EE165 pKa = 5.45FVDD168 pKa = 4.03DD169 pKa = 4.16DD170 pKa = 4.42CVDD173 pKa = 3.88RR174 pKa = 11.84PHH176 pKa = 7.04TDD178 pKa = 2.84SDD180 pKa = 4.2CALEE184 pKa = 4.75LDD186 pKa = 4.98TIITWGIDD194 pKa = 3.38FAGDD198 pKa = 3.75EE199 pKa = 4.46DD200 pKa = 4.11WLKK203 pKa = 11.38ASVDD207 pKa = 3.82TSSGPHH213 pKa = 4.93YY214 pKa = 10.28QFSVANSDD222 pKa = 3.72SQDD225 pKa = 2.99VSLEE229 pKa = 3.74LRR231 pKa = 11.84AQDD234 pKa = 3.97GNLHH238 pKa = 6.22IADD241 pKa = 4.41EE242 pKa = 4.78GADD245 pKa = 3.58EE246 pKa = 4.36ATLTFTTTDD255 pKa = 3.32DD256 pKa = 3.7SNYY259 pKa = 9.01FAAVSFLSSEE269 pKa = 4.21TGDD272 pKa = 4.15FSATYY277 pKa = 8.76TNYY280 pKa = 10.79SGGCPSNPLYY290 pKa = 10.3PSALDD295 pKa = 3.47YY296 pKa = 11.65SNYY299 pKa = 8.64STDD302 pKa = 3.62YY303 pKa = 9.79QKK305 pKa = 11.17CGAEE309 pKa = 4.15APGPDD314 pKa = 3.97AGSATLSGSIEE325 pKa = 4.2PVGDD329 pKa = 3.59TDD331 pKa = 3.33WFKK334 pKa = 10.49ITGMQADD341 pKa = 3.41QSYY344 pKa = 10.42QLDD347 pKa = 4.18LTSGTDD353 pKa = 3.67GLPDD357 pKa = 4.32PLLRR361 pKa = 11.84IYY363 pKa = 10.56DD364 pKa = 3.7AEE366 pKa = 4.32GNEE369 pKa = 4.78LAVDD373 pKa = 3.83DD374 pKa = 5.39DD375 pKa = 4.67GGSKK379 pKa = 10.41EE380 pKa = 4.06DD381 pKa = 3.57AQLGFSPPADD391 pKa = 3.2GDD393 pKa = 3.99YY394 pKa = 10.49FASAEE399 pKa = 4.26SSPKK403 pKa = 9.69DD404 pKa = 3.67DD405 pKa = 3.54QRR407 pKa = 11.84GGYY410 pKa = 8.31SLEE413 pKa = 3.87VANYY417 pKa = 10.1SDD419 pKa = 4.38RR420 pKa = 11.84DD421 pKa = 3.97CAGSNATDD429 pKa = 3.65CGIKK433 pKa = 10.07IGDD436 pKa = 3.85TLVADD441 pKa = 5.75GSTSWDD447 pKa = 3.7YY448 pKa = 10.92EE449 pKa = 4.25VHH451 pKa = 6.65VSGDD455 pKa = 3.72SDD457 pKa = 3.42WFAVSGMTAGEE468 pKa = 4.27SYY470 pKa = 10.79KK471 pKa = 10.7FEE473 pKa = 4.46LRR475 pKa = 11.84NTDD478 pKa = 5.02GILQDD483 pKa = 4.61PYY485 pKa = 11.56LSVYY489 pKa = 10.43DD490 pKa = 3.99SNGEE494 pKa = 3.94EE495 pKa = 4.24LRR497 pKa = 11.84SSGLSKK503 pKa = 11.07DD504 pKa = 3.44PAQLGFTPPDD514 pKa = 3.49NGDD517 pKa = 2.94YY518 pKa = 10.37WIAAEE523 pKa = 4.9SGTEE527 pKa = 3.92GDD529 pKa = 3.13TGAYY533 pKa = 8.19EE534 pKa = 4.2LSAGKK539 pKa = 10.48FNDD542 pKa = 3.65DD543 pKa = 4.17HH544 pKa = 7.3ADD546 pKa = 3.8CATSACSPGTLPVDD560 pKa = 4.13DD561 pKa = 4.62SANGEE566 pKa = 4.27LEE568 pKa = 4.33VPGDD572 pKa = 3.84RR573 pKa = 11.84DD574 pKa = 3.18WFKK577 pKa = 11.8VDD579 pKa = 4.48LEE581 pKa = 4.36SGNYY585 pKa = 7.15YY586 pKa = 9.84TFTLSGDD593 pKa = 3.81TLGAPHH599 pKa = 6.21VRR601 pKa = 11.84LRR603 pKa = 11.84TDD605 pKa = 3.63DD606 pKa = 3.95GSILASQAASPDD618 pKa = 3.31SSEE621 pKa = 4.67AKK623 pKa = 8.86VTHH626 pKa = 6.35EE627 pKa = 4.24PGANQTVYY635 pKa = 11.11LEE637 pKa = 4.33AASVEE642 pKa = 4.25DD643 pKa = 4.64EE644 pKa = 4.34SAGTYY649 pKa = 7.63TVSATAQDD657 pKa = 3.98LSGGTGGGSSDD668 pKa = 3.5GGGSGGGGGCALAAEE683 pKa = 4.73PQPLDD688 pKa = 3.25PTLPLLAGLGLAYY701 pKa = 10.61LLRR704 pKa = 11.84RR705 pKa = 11.84GRR707 pKa = 11.84PRR709 pKa = 11.84AA710 pKa = 3.4
Molecular weight: 74.28 kDa
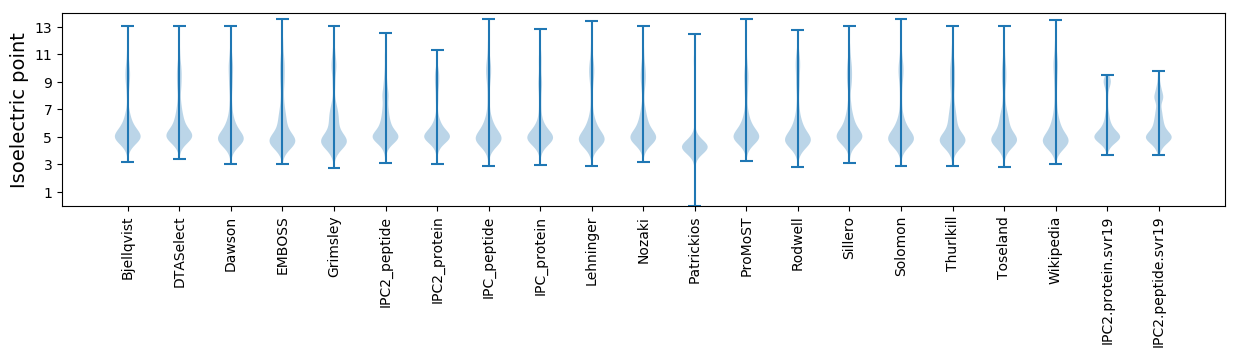
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G5ABI4|A0A1G5ABI4_9GAMM Uncharacterized protein OS=Thiohalorhabdus denitrificans OX=381306 GN=SAMN05661077_0219 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.42HH3 pKa = 5.45TFQPSNIRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.74GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 9.98GGKK28 pKa = 9.08RR29 pKa = 11.84VLKK32 pKa = 10.22RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.84GRR39 pKa = 11.84KK40 pKa = 9.02RR41 pKa = 11.84LTPP44 pKa = 3.95
MM1 pKa = 7.45KK2 pKa = 9.42HH3 pKa = 5.45TFQPSNIRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.74GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 9.98GGKK28 pKa = 9.08RR29 pKa = 11.84VLKK32 pKa = 10.22RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.84GRR39 pKa = 11.84KK40 pKa = 9.02RR41 pKa = 11.84LTPP44 pKa = 3.95
Molecular weight: 5.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
850307 |
33 |
2529 |
315.9 |
34.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.507 ± 0.065 | 0.754 ± 0.014 |
5.612 ± 0.039 | 7.946 ± 0.056 |
3.369 ± 0.028 | 9.454 ± 0.046 |
2.264 ± 0.021 | 3.646 ± 0.034 |
2.393 ± 0.031 | 10.902 ± 0.07 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.118 ± 0.021 | 2.216 ± 0.027 |
5.643 ± 0.034 | 3.24 ± 0.034 |
8.173 ± 0.056 | 4.418 ± 0.032 |
4.765 ± 0.028 | 7.692 ± 0.043 |
1.467 ± 0.021 | 2.421 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
