
Chaetoceros protobacilladnavirus 2 (Chaetoceros sp. DNA virus 7)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Baphyvirales; Bacilladnaviridae; Protobacilladnavirus
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
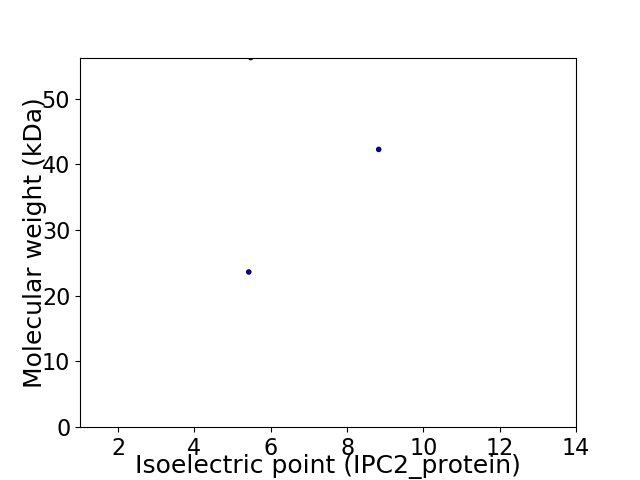
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|W6JIC6|CAPSD_CPBDV Capsid protein OS=Chaetoceros protobacilladnavirus 2 OX=2170141 PE=3 SV=1
MM1 pKa = 6.99NVKK4 pKa = 9.96GASDD8 pKa = 3.66KK9 pKa = 11.47AQLAMQAEE17 pKa = 4.67WEE19 pKa = 4.28EE20 pKa = 4.07VLAPEE25 pKa = 4.5GALAVEE31 pKa = 4.71EE32 pKa = 4.33ASSVLKK38 pKa = 10.77ISDD41 pKa = 3.59DD42 pKa = 3.49EE43 pKa = 4.26RR44 pKa = 11.84RR45 pKa = 11.84SYY47 pKa = 10.69AAYY50 pKa = 9.14IDD52 pKa = 4.99NIIQEE57 pKa = 4.14NEE59 pKa = 3.26IDD61 pKa = 3.76VVYY64 pKa = 10.4GHH66 pKa = 6.37SRR68 pKa = 11.84GAAIASEE75 pKa = 4.63LEE77 pKa = 3.89SDD79 pKa = 3.98VQIIGLDD86 pKa = 3.38GAMVIANDD94 pKa = 3.34QANFLNIRR102 pKa = 11.84QDD104 pKa = 3.25DD105 pKa = 4.15SEE107 pKa = 5.87GYY109 pKa = 10.86GFDD112 pKa = 3.29RR113 pKa = 11.84TIAGPYY119 pKa = 8.47EE120 pKa = 3.68NTVIVKK126 pKa = 10.05GGAFHH131 pKa = 7.27KK132 pKa = 10.57VAVPEE137 pKa = 5.08GYY139 pKa = 8.05HH140 pKa = 4.83TKK142 pKa = 10.3KK143 pKa = 10.53PKK145 pKa = 10.29ASQEE149 pKa = 3.79ALEE152 pKa = 3.94RR153 pKa = 11.84AKK155 pKa = 10.78ARR157 pKa = 11.84KK158 pKa = 9.06HH159 pKa = 4.43NRR161 pKa = 11.84ARR163 pKa = 11.84HH164 pKa = 4.71IARR167 pKa = 11.84AIDD170 pKa = 3.44RR171 pKa = 11.84LTARR175 pKa = 11.84KK176 pKa = 7.59TDD178 pKa = 3.3KK179 pKa = 10.65EE180 pKa = 4.01LEE182 pKa = 4.08EE183 pKa = 4.37IYY185 pKa = 10.04WRR187 pKa = 11.84EE188 pKa = 3.36RR189 pKa = 11.84RR190 pKa = 11.84KK191 pKa = 10.35KK192 pKa = 10.34NKK194 pKa = 10.03NEE196 pKa = 3.57FSKK199 pKa = 11.3ALIEE203 pKa = 4.94FIEE206 pKa = 4.29EE207 pKa = 3.8QLLL210 pKa = 3.73
MM1 pKa = 6.99NVKK4 pKa = 9.96GASDD8 pKa = 3.66KK9 pKa = 11.47AQLAMQAEE17 pKa = 4.67WEE19 pKa = 4.28EE20 pKa = 4.07VLAPEE25 pKa = 4.5GALAVEE31 pKa = 4.71EE32 pKa = 4.33ASSVLKK38 pKa = 10.77ISDD41 pKa = 3.59DD42 pKa = 3.49EE43 pKa = 4.26RR44 pKa = 11.84RR45 pKa = 11.84SYY47 pKa = 10.69AAYY50 pKa = 9.14IDD52 pKa = 4.99NIIQEE57 pKa = 4.14NEE59 pKa = 3.26IDD61 pKa = 3.76VVYY64 pKa = 10.4GHH66 pKa = 6.37SRR68 pKa = 11.84GAAIASEE75 pKa = 4.63LEE77 pKa = 3.89SDD79 pKa = 3.98VQIIGLDD86 pKa = 3.38GAMVIANDD94 pKa = 3.34QANFLNIRR102 pKa = 11.84QDD104 pKa = 3.25DD105 pKa = 4.15SEE107 pKa = 5.87GYY109 pKa = 10.86GFDD112 pKa = 3.29RR113 pKa = 11.84TIAGPYY119 pKa = 8.47EE120 pKa = 3.68NTVIVKK126 pKa = 10.05GGAFHH131 pKa = 7.27KK132 pKa = 10.57VAVPEE137 pKa = 5.08GYY139 pKa = 8.05HH140 pKa = 4.83TKK142 pKa = 10.3KK143 pKa = 10.53PKK145 pKa = 10.29ASQEE149 pKa = 3.79ALEE152 pKa = 3.94RR153 pKa = 11.84AKK155 pKa = 10.78ARR157 pKa = 11.84KK158 pKa = 9.06HH159 pKa = 4.43NRR161 pKa = 11.84ARR163 pKa = 11.84HH164 pKa = 4.71IARR167 pKa = 11.84AIDD170 pKa = 3.44RR171 pKa = 11.84LTARR175 pKa = 11.84KK176 pKa = 7.59TDD178 pKa = 3.3KK179 pKa = 10.65EE180 pKa = 4.01LEE182 pKa = 4.08EE183 pKa = 4.37IYY185 pKa = 10.04WRR187 pKa = 11.84EE188 pKa = 3.36RR189 pKa = 11.84RR190 pKa = 11.84KK191 pKa = 10.35KK192 pKa = 10.34NKK194 pKa = 10.03NEE196 pKa = 3.57FSKK199 pKa = 11.3ALIEE203 pKa = 4.94FIEE206 pKa = 4.29EE207 pKa = 3.8QLLL210 pKa = 3.73
Molecular weight: 23.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|W6JIC6|CAPSD_CPBDV Capsid protein OS=Chaetoceros protobacilladnavirus 2 OX=2170141 PE=3 SV=1
MM1 pKa = 7.61ARR3 pKa = 11.84TKK5 pKa = 10.62SKK7 pKa = 9.63PRR9 pKa = 11.84KK10 pKa = 7.43RR11 pKa = 11.84TTVRR15 pKa = 11.84KK16 pKa = 9.5ARR18 pKa = 11.84RR19 pKa = 11.84SVKK22 pKa = 10.16RR23 pKa = 11.84RR24 pKa = 11.84TTTKK28 pKa = 7.98GTKK31 pKa = 9.6RR32 pKa = 11.84KK33 pKa = 7.41TAGDD37 pKa = 3.84PVTKK41 pKa = 10.23AKK43 pKa = 10.48RR44 pKa = 11.84GVATSSPFAAHH55 pKa = 6.28HH56 pKa = 5.77AVRR59 pKa = 11.84MNPFSGATTQPKK71 pKa = 10.02IPDD74 pKa = 3.57GGFTSSLSRR83 pKa = 11.84RR84 pKa = 11.84LQNVVEE90 pKa = 4.3VTNASNEE97 pKa = 4.48GIMHH101 pKa = 7.19LVMAPTMGVPICVTHH116 pKa = 4.7TTEE119 pKa = 4.41GASTRR124 pKa = 11.84SGATLKK130 pKa = 10.58PSYY133 pKa = 10.54LGFLGQGVGLEE144 pKa = 4.31TKK146 pKa = 10.48VGGVIKK152 pKa = 10.2WPIANTDD159 pKa = 3.22TGDD162 pKa = 3.71VINAADD168 pKa = 3.7FAKK171 pKa = 9.96WRR173 pKa = 11.84VVSQGLQITLNNVDD187 pKa = 5.01DD188 pKa = 5.37EE189 pKa = 4.62NDD191 pKa = 3.14GWFEE195 pKa = 3.72AVRR198 pKa = 11.84FNWRR202 pKa = 11.84NDD204 pKa = 3.37NEE206 pKa = 4.91DD207 pKa = 3.14ICLTSLDD214 pKa = 3.91GTDD217 pKa = 3.14TGNVIGAAPNLNGLPLLTANIVEE240 pKa = 4.28MPGYY244 pKa = 10.47KK245 pKa = 9.74SGLLKK250 pKa = 10.47DD251 pKa = 3.32IKK253 pKa = 10.68DD254 pKa = 3.99YY255 pKa = 11.61QFMLHH260 pKa = 6.23PQQTRR265 pKa = 11.84HH266 pKa = 6.45DD267 pKa = 3.91PVEE270 pKa = 3.93ITKK273 pKa = 10.86SIDD276 pKa = 3.47FVGGTDD282 pKa = 4.55LNYY285 pKa = 9.07DD286 pKa = 3.93TQSKK290 pKa = 9.58KK291 pKa = 11.15ANLGDD296 pKa = 3.97SAAGTLLKK304 pKa = 10.59QGLVDD309 pKa = 5.19QNMDD313 pKa = 2.33WMYY316 pKa = 11.18IRR318 pKa = 11.84IHH320 pKa = 6.19PRR322 pKa = 11.84SNTGAAGQTGSKK334 pKa = 9.82LICNYY339 pKa = 9.62IQNLEE344 pKa = 4.22FAFSPDD350 pKa = 3.21SDD352 pKa = 4.04LATYY356 pKa = 7.5MTTNRR361 pKa = 11.84MDD363 pKa = 3.93PKK365 pKa = 10.22SAKK368 pKa = 10.38INDD371 pKa = 3.8QLNNNQDD378 pKa = 3.23AANKK382 pKa = 9.55KK383 pKa = 10.34RR384 pKa = 11.84EE385 pKa = 4.1FNN387 pKa = 3.6
MM1 pKa = 7.61ARR3 pKa = 11.84TKK5 pKa = 10.62SKK7 pKa = 9.63PRR9 pKa = 11.84KK10 pKa = 7.43RR11 pKa = 11.84TTVRR15 pKa = 11.84KK16 pKa = 9.5ARR18 pKa = 11.84RR19 pKa = 11.84SVKK22 pKa = 10.16RR23 pKa = 11.84RR24 pKa = 11.84TTTKK28 pKa = 7.98GTKK31 pKa = 9.6RR32 pKa = 11.84KK33 pKa = 7.41TAGDD37 pKa = 3.84PVTKK41 pKa = 10.23AKK43 pKa = 10.48RR44 pKa = 11.84GVATSSPFAAHH55 pKa = 6.28HH56 pKa = 5.77AVRR59 pKa = 11.84MNPFSGATTQPKK71 pKa = 10.02IPDD74 pKa = 3.57GGFTSSLSRR83 pKa = 11.84RR84 pKa = 11.84LQNVVEE90 pKa = 4.3VTNASNEE97 pKa = 4.48GIMHH101 pKa = 7.19LVMAPTMGVPICVTHH116 pKa = 4.7TTEE119 pKa = 4.41GASTRR124 pKa = 11.84SGATLKK130 pKa = 10.58PSYY133 pKa = 10.54LGFLGQGVGLEE144 pKa = 4.31TKK146 pKa = 10.48VGGVIKK152 pKa = 10.2WPIANTDD159 pKa = 3.22TGDD162 pKa = 3.71VINAADD168 pKa = 3.7FAKK171 pKa = 9.96WRR173 pKa = 11.84VVSQGLQITLNNVDD187 pKa = 5.01DD188 pKa = 5.37EE189 pKa = 4.62NDD191 pKa = 3.14GWFEE195 pKa = 3.72AVRR198 pKa = 11.84FNWRR202 pKa = 11.84NDD204 pKa = 3.37NEE206 pKa = 4.91DD207 pKa = 3.14ICLTSLDD214 pKa = 3.91GTDD217 pKa = 3.14TGNVIGAAPNLNGLPLLTANIVEE240 pKa = 4.28MPGYY244 pKa = 10.47KK245 pKa = 9.74SGLLKK250 pKa = 10.47DD251 pKa = 3.32IKK253 pKa = 10.68DD254 pKa = 3.99YY255 pKa = 11.61QFMLHH260 pKa = 6.23PQQTRR265 pKa = 11.84HH266 pKa = 6.45DD267 pKa = 3.91PVEE270 pKa = 3.93ITKK273 pKa = 10.86SIDD276 pKa = 3.47FVGGTDD282 pKa = 4.55LNYY285 pKa = 9.07DD286 pKa = 3.93TQSKK290 pKa = 9.58KK291 pKa = 11.15ANLGDD296 pKa = 3.97SAAGTLLKK304 pKa = 10.59QGLVDD309 pKa = 5.19QNMDD313 pKa = 2.33WMYY316 pKa = 11.18IRR318 pKa = 11.84IHH320 pKa = 6.19PRR322 pKa = 11.84SNTGAAGQTGSKK334 pKa = 9.82LICNYY339 pKa = 9.62IQNLEE344 pKa = 4.22FAFSPDD350 pKa = 3.21SDD352 pKa = 4.04LATYY356 pKa = 7.5MTTNRR361 pKa = 11.84MDD363 pKa = 3.93PKK365 pKa = 10.22SAKK368 pKa = 10.38INDD371 pKa = 3.8QLNNNQDD378 pKa = 3.23AANKK382 pKa = 9.55KK383 pKa = 10.34RR384 pKa = 11.84EE385 pKa = 4.1FNN387 pKa = 3.6
Molecular weight: 42.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1081 |
210 |
484 |
360.3 |
40.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.048 ± 1.78 | 1.11 ± 0.463 |
6.846 ± 0.252 | 7.216 ± 2.139 |
4.07 ± 0.874 | 6.846 ± 0.79 |
3.053 ± 0.737 | 5.365 ± 0.861 |
7.216 ± 0.145 | 5.828 ± 0.631 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.758 ± 0.514 | 5.92 ± 0.743 |
4.718 ± 0.964 | 4.07 ± 0.153 |
5.92 ± 0.387 | 5.643 ± 0.163 |
6.383 ± 1.764 | 5.273 ± 0.538 |
1.943 ± 0.546 | 2.775 ± 0.452 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
