
Apis mellifera associated microvirus 23
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.62
Get precalculated fractions of proteins
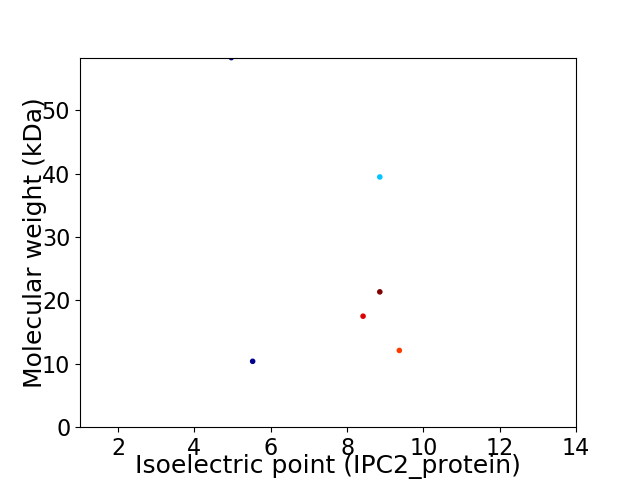
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
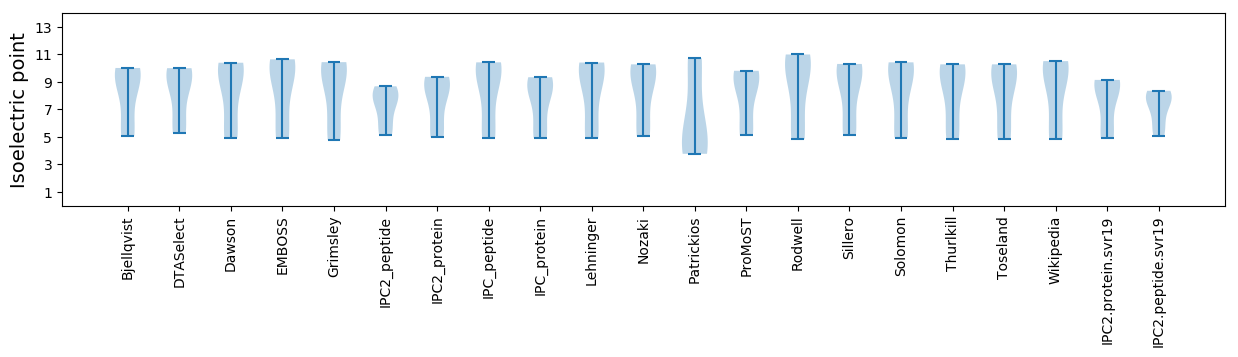
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UTX4|A0A3S8UTX4_9VIRU Internal scaffolding protein OS=Apis mellifera associated microvirus 23 OX=2494751 PE=4 SV=1
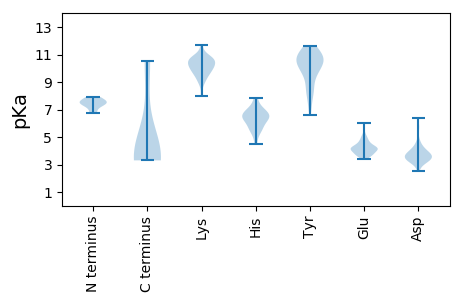
MM1 pKa = 7.4LGSRR5 pKa = 11.84MSQHH9 pKa = 6.3KK10 pKa = 8.96FANVPAVNTARR21 pKa = 11.84SSFDD25 pKa = 2.99RR26 pKa = 11.84SFQIKK31 pKa = 8.78DD32 pKa = 3.42TFDD35 pKa = 3.54FDD37 pKa = 3.92YY38 pKa = 10.84LIPIFVDD45 pKa = 4.73EE46 pKa = 4.88ILPGDD51 pKa = 3.75TCNVRR56 pKa = 11.84MSCFGRR62 pKa = 11.84LATQLVPIMDD72 pKa = 4.07NMYY75 pKa = 9.86IDD77 pKa = 3.75YY78 pKa = 10.82FFFFVPNRR86 pKa = 11.84LVWDD90 pKa = 3.1NWEE93 pKa = 4.33RR94 pKa = 11.84FNGAQDD100 pKa = 4.11DD101 pKa = 4.68PGDD104 pKa = 4.02STDD107 pKa = 3.85FTIPQIEE114 pKa = 4.68FATRR118 pKa = 11.84PQVGDD123 pKa = 3.29IYY125 pKa = 11.43DD126 pKa = 3.81HH127 pKa = 6.93FGLPTGLTNTWYY139 pKa = 10.94CNALPFRR146 pKa = 11.84CYY148 pKa = 11.02SLIWNEE154 pKa = 3.67WFRR157 pKa = 11.84DD158 pKa = 3.62QNLQSSVPVPKK169 pKa = 10.72DD170 pKa = 3.24NGPDD174 pKa = 3.25VYY176 pKa = 11.6ADD178 pKa = 3.56FTLLKK183 pKa = 9.88RR184 pKa = 11.84GKK186 pKa = 9.0RR187 pKa = 11.84HH188 pKa = 7.53DD189 pKa = 4.2YY190 pKa = 8.21FTSCLPWPQKK200 pKa = 11.05GPDD203 pKa = 3.33VTLPLASQAPVKK215 pKa = 10.81SLGVVGNDD223 pKa = 3.2YY224 pKa = 11.44VGVLDD229 pKa = 3.74STNTPRR235 pKa = 11.84RR236 pKa = 11.84LQDD239 pKa = 2.98NGTAIIIAAGSPTASADD256 pKa = 3.4LFADD260 pKa = 4.45LNQATAATINQLRR273 pKa = 11.84EE274 pKa = 3.99AFLMQSLFEE283 pKa = 5.07LDD285 pKa = 3.28ARR287 pKa = 11.84GGTRR291 pKa = 11.84YY292 pKa = 9.98VEE294 pKa = 3.69ILQAHH299 pKa = 6.38FNVTSPDD306 pKa = 3.15FRR308 pKa = 11.84LQRR311 pKa = 11.84PEE313 pKa = 3.69FLGGGSSRR321 pKa = 11.84INSHH325 pKa = 6.57IVPQTSPTSGSDD337 pKa = 3.4PQGQLAAFATTSADD351 pKa = 3.62GIGFTKK357 pKa = 10.61SFVEE361 pKa = 3.76HH362 pKa = 6.81GYY364 pKa = 11.06VIGLACARR372 pKa = 11.84ADD374 pKa = 3.33VTYY377 pKa = 10.54QQGLNRR383 pKa = 11.84MWSRR387 pKa = 11.84STRR390 pKa = 11.84YY391 pKa = 10.5DD392 pKa = 3.47FFWPKK397 pKa = 10.2LQEE400 pKa = 4.22LGEE403 pKa = 4.13QEE405 pKa = 4.18VLNRR409 pKa = 11.84EE410 pKa = 4.12IYY412 pKa = 10.53LQGTADD418 pKa = 3.83DD419 pKa = 4.28EE420 pKa = 5.03AVFGYY425 pKa = 7.37QEE427 pKa = 4.18RR428 pKa = 11.84YY429 pKa = 9.58AEE431 pKa = 4.06YY432 pKa = 9.72RR433 pKa = 11.84YY434 pKa = 10.26KK435 pKa = 10.54PSEE438 pKa = 3.37IRR440 pKa = 11.84GQFRR444 pKa = 11.84STYY447 pKa = 9.06STSLDD452 pKa = 3.35FWHH455 pKa = 6.56MAEE458 pKa = 4.67EE459 pKa = 4.63FGSLPALNDD468 pKa = 3.25TFIQQNTPVDD478 pKa = 3.63RR479 pKa = 11.84AIAIPTEE486 pKa = 3.66PHH488 pKa = 6.48LLFDD492 pKa = 4.82AYY494 pKa = 10.7FKK496 pKa = 9.05YY497 pKa = 8.69THH499 pKa = 6.04VRR501 pKa = 11.84PMMTYY506 pKa = 8.51STPASLGRR514 pKa = 11.84FF515 pKa = 3.67
MM1 pKa = 7.4LGSRR5 pKa = 11.84MSQHH9 pKa = 6.3KK10 pKa = 8.96FANVPAVNTARR21 pKa = 11.84SSFDD25 pKa = 2.99RR26 pKa = 11.84SFQIKK31 pKa = 8.78DD32 pKa = 3.42TFDD35 pKa = 3.54FDD37 pKa = 3.92YY38 pKa = 10.84LIPIFVDD45 pKa = 4.73EE46 pKa = 4.88ILPGDD51 pKa = 3.75TCNVRR56 pKa = 11.84MSCFGRR62 pKa = 11.84LATQLVPIMDD72 pKa = 4.07NMYY75 pKa = 9.86IDD77 pKa = 3.75YY78 pKa = 10.82FFFFVPNRR86 pKa = 11.84LVWDD90 pKa = 3.1NWEE93 pKa = 4.33RR94 pKa = 11.84FNGAQDD100 pKa = 4.11DD101 pKa = 4.68PGDD104 pKa = 4.02STDD107 pKa = 3.85FTIPQIEE114 pKa = 4.68FATRR118 pKa = 11.84PQVGDD123 pKa = 3.29IYY125 pKa = 11.43DD126 pKa = 3.81HH127 pKa = 6.93FGLPTGLTNTWYY139 pKa = 10.94CNALPFRR146 pKa = 11.84CYY148 pKa = 11.02SLIWNEE154 pKa = 3.67WFRR157 pKa = 11.84DD158 pKa = 3.62QNLQSSVPVPKK169 pKa = 10.72DD170 pKa = 3.24NGPDD174 pKa = 3.25VYY176 pKa = 11.6ADD178 pKa = 3.56FTLLKK183 pKa = 9.88RR184 pKa = 11.84GKK186 pKa = 9.0RR187 pKa = 11.84HH188 pKa = 7.53DD189 pKa = 4.2YY190 pKa = 8.21FTSCLPWPQKK200 pKa = 11.05GPDD203 pKa = 3.33VTLPLASQAPVKK215 pKa = 10.81SLGVVGNDD223 pKa = 3.2YY224 pKa = 11.44VGVLDD229 pKa = 3.74STNTPRR235 pKa = 11.84RR236 pKa = 11.84LQDD239 pKa = 2.98NGTAIIIAAGSPTASADD256 pKa = 3.4LFADD260 pKa = 4.45LNQATAATINQLRR273 pKa = 11.84EE274 pKa = 3.99AFLMQSLFEE283 pKa = 5.07LDD285 pKa = 3.28ARR287 pKa = 11.84GGTRR291 pKa = 11.84YY292 pKa = 9.98VEE294 pKa = 3.69ILQAHH299 pKa = 6.38FNVTSPDD306 pKa = 3.15FRR308 pKa = 11.84LQRR311 pKa = 11.84PEE313 pKa = 3.69FLGGGSSRR321 pKa = 11.84INSHH325 pKa = 6.57IVPQTSPTSGSDD337 pKa = 3.4PQGQLAAFATTSADD351 pKa = 3.62GIGFTKK357 pKa = 10.61SFVEE361 pKa = 3.76HH362 pKa = 6.81GYY364 pKa = 11.06VIGLACARR372 pKa = 11.84ADD374 pKa = 3.33VTYY377 pKa = 10.54QQGLNRR383 pKa = 11.84MWSRR387 pKa = 11.84STRR390 pKa = 11.84YY391 pKa = 10.5DD392 pKa = 3.47FFWPKK397 pKa = 10.2LQEE400 pKa = 4.22LGEE403 pKa = 4.13QEE405 pKa = 4.18VLNRR409 pKa = 11.84EE410 pKa = 4.12IYY412 pKa = 10.53LQGTADD418 pKa = 3.83DD419 pKa = 4.28EE420 pKa = 5.03AVFGYY425 pKa = 7.37QEE427 pKa = 4.18RR428 pKa = 11.84YY429 pKa = 9.58AEE431 pKa = 4.06YY432 pKa = 9.72RR433 pKa = 11.84YY434 pKa = 10.26KK435 pKa = 10.54PSEE438 pKa = 3.37IRR440 pKa = 11.84GQFRR444 pKa = 11.84STYY447 pKa = 9.06STSLDD452 pKa = 3.35FWHH455 pKa = 6.56MAEE458 pKa = 4.67EE459 pKa = 4.63FGSLPALNDD468 pKa = 3.25TFIQQNTPVDD478 pKa = 3.63RR479 pKa = 11.84AIAIPTEE486 pKa = 3.66PHH488 pKa = 6.48LLFDD492 pKa = 4.82AYY494 pKa = 10.7FKK496 pKa = 9.05YY497 pKa = 8.69THH499 pKa = 6.04VRR501 pKa = 11.84PMMTYY506 pKa = 8.51STPASLGRR514 pKa = 11.84FF515 pKa = 3.67
Molecular weight: 58.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UTW3|A0A3S8UTW3_9VIRU Major capsid protein OS=Apis mellifera associated microvirus 23 OX=2494751 PE=3 SV=1
MM1 pKa = 7.41EE2 pKa = 5.03GVRR5 pKa = 11.84EE6 pKa = 4.78AIACPWKK13 pKa = 10.5VKK15 pKa = 10.36KK16 pKa = 10.41IYY18 pKa = 9.63LQKK21 pKa = 10.96NLYY24 pKa = 10.06KK25 pKa = 10.53LACNQQIAGNEE36 pKa = 3.83NSASMRR42 pKa = 11.84RR43 pKa = 11.84KK44 pKa = 10.11AVMQRR49 pKa = 11.84AQYY52 pKa = 9.57EE53 pKa = 4.16NWKK56 pKa = 10.08KK57 pKa = 9.89RR58 pKa = 11.84VRR60 pKa = 11.84RR61 pKa = 11.84EE62 pKa = 3.76FKK64 pKa = 10.63KK65 pKa = 10.88LAGEE69 pKa = 4.28TKK71 pKa = 10.23KK72 pKa = 11.02VEE74 pKa = 4.1GLGWSQYY81 pKa = 9.53LDD83 pKa = 3.35KK84 pKa = 10.61RR85 pKa = 11.84VKK87 pKa = 10.42EE88 pKa = 3.64IGFRR92 pKa = 11.84SINHH96 pKa = 6.48LCAEE100 pKa = 4.81LSQQ103 pKa = 3.75
MM1 pKa = 7.41EE2 pKa = 5.03GVRR5 pKa = 11.84EE6 pKa = 4.78AIACPWKK13 pKa = 10.5VKK15 pKa = 10.36KK16 pKa = 10.41IYY18 pKa = 9.63LQKK21 pKa = 10.96NLYY24 pKa = 10.06KK25 pKa = 10.53LACNQQIAGNEE36 pKa = 3.83NSASMRR42 pKa = 11.84RR43 pKa = 11.84KK44 pKa = 10.11AVMQRR49 pKa = 11.84AQYY52 pKa = 9.57EE53 pKa = 4.16NWKK56 pKa = 10.08KK57 pKa = 9.89RR58 pKa = 11.84VRR60 pKa = 11.84RR61 pKa = 11.84EE62 pKa = 3.76FKK64 pKa = 10.63KK65 pKa = 10.88LAGEE69 pKa = 4.28TKK71 pKa = 10.23KK72 pKa = 11.02VEE74 pKa = 4.1GLGWSQYY81 pKa = 9.53LDD83 pKa = 3.35KK84 pKa = 10.61RR85 pKa = 11.84VKK87 pKa = 10.42EE88 pKa = 3.64IGFRR92 pKa = 11.84SINHH96 pKa = 6.48LCAEE100 pKa = 4.81LSQQ103 pKa = 3.75
Molecular weight: 12.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1398 |
90 |
515 |
233.0 |
26.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.369 ± 1.394 | 1.359 ± 0.486 |
6.295 ± 0.801 | 5.508 ± 0.739 |
5.079 ± 1.05 | 6.009 ± 0.623 |
1.86 ± 0.41 | 4.649 ± 0.43 |
7.01 ± 2.162 | 7.296 ± 0.458 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.289 ± 0.427 | 4.864 ± 0.989 |
5.007 ± 0.784 | 5.222 ± 0.298 |
6.652 ± 0.563 | 6.652 ± 0.577 |
5.365 ± 0.934 | 4.936 ± 0.519 |
1.502 ± 0.457 | 4.077 ± 0.722 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
