
Capuchin monkey hepatitis B virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Blubervirales; Hepadnaviridae; Orthohepadnavirus
Average proteome isoelectric point is 8.15
Get precalculated fractions of proteins
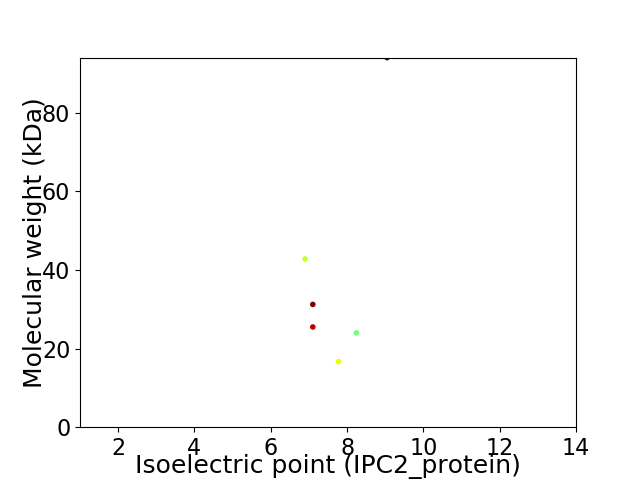
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
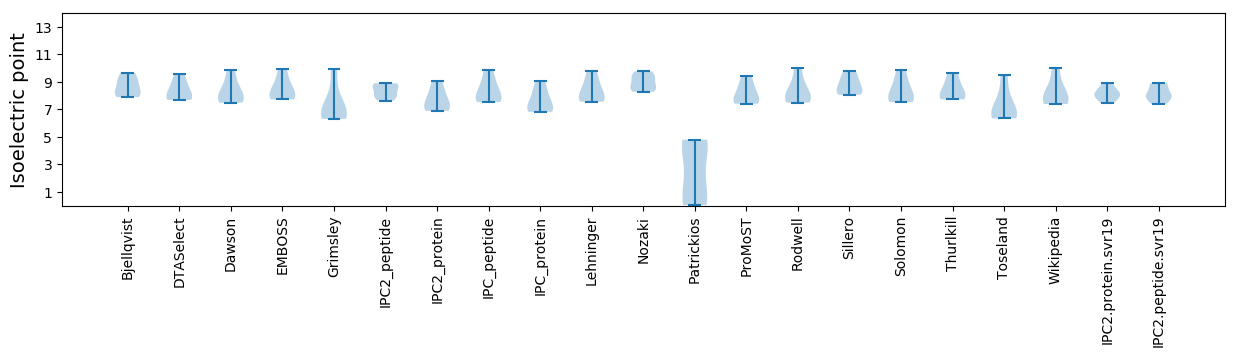
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2R4KZU8|A0A2R4KZU8_9HEPA Protein X OS=Capuchin monkey hepatitis B virus OX=2163996 GN=X PE=3 SV=1
MM1 pKa = 7.71GLNQSTINPLGFFPSHH17 pKa = 6.03QLDD20 pKa = 3.75PLFKK24 pKa = 10.35ANSPAADD31 pKa = 3.47WDD33 pKa = 4.12QNPHH37 pKa = 6.38KK38 pKa = 10.57DD39 pKa = 3.48PWPSAHH45 pKa = 6.29EE46 pKa = 4.3VPVGAFGPGLVPPHH60 pKa = 6.62GGLLGWNPQSQGQVTLLPKK79 pKa = 10.49DD80 pKa = 4.01PPAQLPNRR88 pKa = 11.84NSARR92 pKa = 11.84VPTPITPPLRR102 pKa = 11.84DD103 pKa = 3.51THH105 pKa = 5.95PQAMTWNISNFQKK118 pKa = 10.81ALQDD122 pKa = 3.33PRR124 pKa = 11.84VRR126 pKa = 11.84GLYY129 pKa = 9.96FPAGGSSSGIPTPAPTTASTISSLFATTGDD159 pKa = 3.18PVEE162 pKa = 4.21NMEE165 pKa = 4.96NITSGFLGPLLALQAVFFLLTKK187 pKa = 10.24ILTIPQSLDD196 pKa = 3.56SLWTSLNFLGEE207 pKa = 4.28TPACPGRR214 pKa = 11.84NLQSPTCSHH223 pKa = 6.69SPTSCPQTCPGYY235 pKa = 10.61RR236 pKa = 11.84WMCLRR241 pKa = 11.84RR242 pKa = 11.84FIIFLFIPLLCLIFLLVLLDD262 pKa = 3.96YY263 pKa = 11.31QGMLPVCPLLPGTTTTTTTGPCKK286 pKa = 9.66TCTPIAPGSSMYY298 pKa = 10.25PSCCCTKK305 pKa = 9.49PTDD308 pKa = 4.27GNCTCIPIPSSWAFARR324 pKa = 11.84FLWDD328 pKa = 2.94WASVRR333 pKa = 11.84FSWLSLLAPFVQWFVGLSPTAWLLVIWMIWFWGPSLYY370 pKa = 10.49TILSPFFPLLPVFFCLWVYY389 pKa = 10.5II390 pKa = 4.23
MM1 pKa = 7.71GLNQSTINPLGFFPSHH17 pKa = 6.03QLDD20 pKa = 3.75PLFKK24 pKa = 10.35ANSPAADD31 pKa = 3.47WDD33 pKa = 4.12QNPHH37 pKa = 6.38KK38 pKa = 10.57DD39 pKa = 3.48PWPSAHH45 pKa = 6.29EE46 pKa = 4.3VPVGAFGPGLVPPHH60 pKa = 6.62GGLLGWNPQSQGQVTLLPKK79 pKa = 10.49DD80 pKa = 4.01PPAQLPNRR88 pKa = 11.84NSARR92 pKa = 11.84VPTPITPPLRR102 pKa = 11.84DD103 pKa = 3.51THH105 pKa = 5.95PQAMTWNISNFQKK118 pKa = 10.81ALQDD122 pKa = 3.33PRR124 pKa = 11.84VRR126 pKa = 11.84GLYY129 pKa = 9.96FPAGGSSSGIPTPAPTTASTISSLFATTGDD159 pKa = 3.18PVEE162 pKa = 4.21NMEE165 pKa = 4.96NITSGFLGPLLALQAVFFLLTKK187 pKa = 10.24ILTIPQSLDD196 pKa = 3.56SLWTSLNFLGEE207 pKa = 4.28TPACPGRR214 pKa = 11.84NLQSPTCSHH223 pKa = 6.69SPTSCPQTCPGYY235 pKa = 10.61RR236 pKa = 11.84WMCLRR241 pKa = 11.84RR242 pKa = 11.84FIIFLFIPLLCLIFLLVLLDD262 pKa = 3.96YY263 pKa = 11.31QGMLPVCPLLPGTTTTTTTGPCKK286 pKa = 9.66TCTPIAPGSSMYY298 pKa = 10.25PSCCCTKK305 pKa = 9.49PTDD308 pKa = 4.27GNCTCIPIPSSWAFARR324 pKa = 11.84FLWDD328 pKa = 2.94WASVRR333 pKa = 11.84FSWLSLLAPFVQWFVGLSPTAWLLVIWMIWFWGPSLYY370 pKa = 10.49TILSPFFPLLPVFFCLWVYY389 pKa = 10.5II390 pKa = 4.23
Molecular weight: 42.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R4QN08|A0A2R4QN08_9HEPA Middle surface antigen OS=Capuchin monkey hepatitis B virus OX=2163996 PE=4 SV=1
MM1 pKa = 7.8PLSCQLFRR9 pKa = 11.84KK10 pKa = 9.82LLLLEE15 pKa = 4.89GDD17 pKa = 3.51QGPLEE22 pKa = 4.34EE23 pKa = 4.74EE24 pKa = 4.39LPRR27 pKa = 11.84LADD30 pKa = 3.38EE31 pKa = 4.61DD32 pKa = 4.03LNRR35 pKa = 11.84RR36 pKa = 11.84VAEE39 pKa = 4.31DD40 pKa = 4.58LNLQLPPVSIPWTHH54 pKa = 5.68TVGEE58 pKa = 4.34FTGLYY63 pKa = 9.97SVTPLRR69 pKa = 11.84YY70 pKa = 9.03NPEE73 pKa = 3.45WKK75 pKa = 10.09IPEE78 pKa = 4.28FPNIHH83 pKa = 6.73LKK85 pKa = 10.04QDD87 pKa = 3.38IIKK90 pKa = 10.32RR91 pKa = 11.84CTDD94 pKa = 3.64FVGPLNANEE103 pKa = 3.9TRR105 pKa = 11.84RR106 pKa = 11.84LKK108 pKa = 10.86LVLPARR114 pKa = 11.84FYY116 pKa = 11.11PKK118 pKa = 8.17VTKK121 pKa = 10.3YY122 pKa = 10.46FPLEE126 pKa = 4.06KK127 pKa = 9.93GIKK130 pKa = 8.49PHH132 pKa = 7.02YY133 pKa = 8.48PDD135 pKa = 3.51QVVNHH140 pKa = 5.4YY141 pKa = 10.63FKK143 pKa = 10.87VQEE146 pKa = 4.11YY147 pKa = 10.52LHH149 pKa = 6.05TLWKK153 pKa = 10.61SGILYY158 pKa = 10.13KK159 pKa = 10.5RR160 pKa = 11.84EE161 pKa = 4.21TTHH164 pKa = 6.06SASFFGSPYY173 pKa = 8.4TWEE176 pKa = 4.26HH177 pKa = 5.18QLQHH181 pKa = 5.04GTKK184 pKa = 9.22PVYY187 pKa = 9.64HH188 pKa = 6.34QSSGVLPQSSARR200 pKa = 11.84PLVQSEE206 pKa = 4.54FTSSRR211 pKa = 11.84LGSKK215 pKa = 10.16SSQRR219 pKa = 11.84PLAFSSRR226 pKa = 11.84SACRR230 pKa = 11.84GLWTRR235 pKa = 11.84PSSSSRR241 pKa = 11.84RR242 pKa = 11.84FARR245 pKa = 11.84LEE247 pKa = 3.91SSVTGSSHH255 pKa = 6.62PVAKK259 pKa = 10.38RR260 pKa = 11.84SACSTAQSQLSQSTHH275 pKa = 4.23SHH277 pKa = 4.56NTTTKK282 pKa = 8.24GHH284 pKa = 6.52SSPSHH289 pKa = 5.72DD290 pKa = 4.55LEE292 pKa = 4.07YY293 pKa = 11.15LKK295 pKa = 10.91LSEE298 pKa = 4.33GSSRR302 pKa = 11.84SSSQGAVFPCWWLQFRR318 pKa = 11.84DD319 pKa = 4.08SNPCSDD325 pKa = 3.96YY326 pKa = 10.79CLHH329 pKa = 6.93HH330 pKa = 6.62IVSLRR335 pKa = 11.84DD336 pKa = 3.06NWGPCGEE343 pKa = 4.61HH344 pKa = 6.65GEE346 pKa = 4.06HH347 pKa = 7.36HH348 pKa = 6.08IRR350 pKa = 11.84IPRR353 pKa = 11.84TPARR357 pKa = 11.84ITGGVFLVDD366 pKa = 4.0KK367 pKa = 10.39NPHH370 pKa = 5.83NSSEE374 pKa = 4.07SRR376 pKa = 11.84LVVDD380 pKa = 5.53FSQFSRR386 pKa = 11.84GDD388 pKa = 3.55TRR390 pKa = 11.84VSWPKK395 pKa = 9.99FAVPNLQSLTNLLSSDD411 pKa = 4.31LSWLSLDD418 pKa = 3.48VSAAFYY424 pKa = 10.35HH425 pKa = 6.61LPLHH429 pKa = 6.16PAVMPHH435 pKa = 6.65LLVGTAGLSRR445 pKa = 11.84YY446 pKa = 7.18VARR449 pKa = 11.84LSSPARR455 pKa = 11.84NNNHH459 pKa = 5.34HH460 pKa = 6.31HH461 pKa = 6.49HH462 pKa = 6.52RR463 pKa = 11.84TLQDD467 pKa = 2.99LHH469 pKa = 6.29AHH471 pKa = 5.84CTRR474 pKa = 11.84EE475 pKa = 4.18LYY477 pKa = 10.68VSLMLLYY484 pKa = 8.84QTYY487 pKa = 9.1GWKK490 pKa = 10.04LHH492 pKa = 6.48LYY494 pKa = 8.45SHH496 pKa = 7.36PIVLGFRR503 pKa = 11.84KK504 pKa = 9.99VPMGLGLSPFLLAQFTSAICSVVRR528 pKa = 11.84RR529 pKa = 11.84AFPHH533 pKa = 6.38CMAFSYY539 pKa = 10.53MDD541 pKa = 4.5DD542 pKa = 3.65LVLGAKK548 pKa = 9.36SVHH551 pKa = 6.13HH552 pKa = 6.53LEE554 pKa = 4.43SLFSAITSFLLSLGIHH570 pKa = 6.68LNPEE574 pKa = 3.65KK575 pKa = 10.04TKK577 pKa = 10.52RR578 pKa = 11.84WGKK581 pKa = 9.39SLQFMGYY588 pKa = 10.21VIGAWGSLPQDD599 pKa = 4.11HH600 pKa = 6.82IRR602 pKa = 11.84AKK604 pKa = 9.95IQRR607 pKa = 11.84CFRR610 pKa = 11.84TLPCNRR616 pKa = 11.84PIDD619 pKa = 3.55WKK621 pKa = 8.96VCQRR625 pKa = 11.84IVGLLGFAAPFTQCGYY641 pKa = 8.88PALMPIYY648 pKa = 10.04HH649 pKa = 7.4AIKK652 pKa = 10.19LKK654 pKa = 10.31QAFSFSLTYY663 pKa = 10.44KK664 pKa = 10.77AFLRR668 pKa = 11.84DD669 pKa = 3.4QYY671 pKa = 11.04LTLHH675 pKa = 6.18PVARR679 pKa = 11.84QRR681 pKa = 11.84PGPCQVFADD690 pKa = 4.34ATPTGWGLVCGQQRR704 pKa = 11.84MRR706 pKa = 11.84GDD708 pKa = 3.89FVSPLPIHH716 pKa = 5.41TAEE719 pKa = 4.49LLAACFARR727 pKa = 11.84CWTGAKK733 pKa = 9.96HH734 pKa = 6.11IATDD738 pKa = 3.49NSVVLSRR745 pKa = 11.84KK746 pKa = 7.43YY747 pKa = 9.41TSFPWLLGCAANWILRR763 pKa = 11.84GTCFVYY769 pKa = 10.73VPSQQNPADD778 pKa = 4.03DD779 pKa = 4.24PSRR782 pKa = 11.84GRR784 pKa = 11.84LGLLRR789 pKa = 11.84PLPRR793 pKa = 11.84LHH795 pKa = 6.93FLPSTGRR802 pKa = 11.84TSLYY806 pKa = 10.51ADD808 pKa = 3.69SPPVPFHH815 pKa = 6.63RR816 pKa = 11.84PARR819 pKa = 11.84VHH821 pKa = 6.17FASPLHH827 pKa = 6.31DD828 pKa = 3.21AWRR831 pKa = 11.84PPP833 pKa = 3.23
MM1 pKa = 7.8PLSCQLFRR9 pKa = 11.84KK10 pKa = 9.82LLLLEE15 pKa = 4.89GDD17 pKa = 3.51QGPLEE22 pKa = 4.34EE23 pKa = 4.74EE24 pKa = 4.39LPRR27 pKa = 11.84LADD30 pKa = 3.38EE31 pKa = 4.61DD32 pKa = 4.03LNRR35 pKa = 11.84RR36 pKa = 11.84VAEE39 pKa = 4.31DD40 pKa = 4.58LNLQLPPVSIPWTHH54 pKa = 5.68TVGEE58 pKa = 4.34FTGLYY63 pKa = 9.97SVTPLRR69 pKa = 11.84YY70 pKa = 9.03NPEE73 pKa = 3.45WKK75 pKa = 10.09IPEE78 pKa = 4.28FPNIHH83 pKa = 6.73LKK85 pKa = 10.04QDD87 pKa = 3.38IIKK90 pKa = 10.32RR91 pKa = 11.84CTDD94 pKa = 3.64FVGPLNANEE103 pKa = 3.9TRR105 pKa = 11.84RR106 pKa = 11.84LKK108 pKa = 10.86LVLPARR114 pKa = 11.84FYY116 pKa = 11.11PKK118 pKa = 8.17VTKK121 pKa = 10.3YY122 pKa = 10.46FPLEE126 pKa = 4.06KK127 pKa = 9.93GIKK130 pKa = 8.49PHH132 pKa = 7.02YY133 pKa = 8.48PDD135 pKa = 3.51QVVNHH140 pKa = 5.4YY141 pKa = 10.63FKK143 pKa = 10.87VQEE146 pKa = 4.11YY147 pKa = 10.52LHH149 pKa = 6.05TLWKK153 pKa = 10.61SGILYY158 pKa = 10.13KK159 pKa = 10.5RR160 pKa = 11.84EE161 pKa = 4.21TTHH164 pKa = 6.06SASFFGSPYY173 pKa = 8.4TWEE176 pKa = 4.26HH177 pKa = 5.18QLQHH181 pKa = 5.04GTKK184 pKa = 9.22PVYY187 pKa = 9.64HH188 pKa = 6.34QSSGVLPQSSARR200 pKa = 11.84PLVQSEE206 pKa = 4.54FTSSRR211 pKa = 11.84LGSKK215 pKa = 10.16SSQRR219 pKa = 11.84PLAFSSRR226 pKa = 11.84SACRR230 pKa = 11.84GLWTRR235 pKa = 11.84PSSSSRR241 pKa = 11.84RR242 pKa = 11.84FARR245 pKa = 11.84LEE247 pKa = 3.91SSVTGSSHH255 pKa = 6.62PVAKK259 pKa = 10.38RR260 pKa = 11.84SACSTAQSQLSQSTHH275 pKa = 4.23SHH277 pKa = 4.56NTTTKK282 pKa = 8.24GHH284 pKa = 6.52SSPSHH289 pKa = 5.72DD290 pKa = 4.55LEE292 pKa = 4.07YY293 pKa = 11.15LKK295 pKa = 10.91LSEE298 pKa = 4.33GSSRR302 pKa = 11.84SSSQGAVFPCWWLQFRR318 pKa = 11.84DD319 pKa = 4.08SNPCSDD325 pKa = 3.96YY326 pKa = 10.79CLHH329 pKa = 6.93HH330 pKa = 6.62IVSLRR335 pKa = 11.84DD336 pKa = 3.06NWGPCGEE343 pKa = 4.61HH344 pKa = 6.65GEE346 pKa = 4.06HH347 pKa = 7.36HH348 pKa = 6.08IRR350 pKa = 11.84IPRR353 pKa = 11.84TPARR357 pKa = 11.84ITGGVFLVDD366 pKa = 4.0KK367 pKa = 10.39NPHH370 pKa = 5.83NSSEE374 pKa = 4.07SRR376 pKa = 11.84LVVDD380 pKa = 5.53FSQFSRR386 pKa = 11.84GDD388 pKa = 3.55TRR390 pKa = 11.84VSWPKK395 pKa = 9.99FAVPNLQSLTNLLSSDD411 pKa = 4.31LSWLSLDD418 pKa = 3.48VSAAFYY424 pKa = 10.35HH425 pKa = 6.61LPLHH429 pKa = 6.16PAVMPHH435 pKa = 6.65LLVGTAGLSRR445 pKa = 11.84YY446 pKa = 7.18VARR449 pKa = 11.84LSSPARR455 pKa = 11.84NNNHH459 pKa = 5.34HH460 pKa = 6.31HH461 pKa = 6.49HH462 pKa = 6.52RR463 pKa = 11.84TLQDD467 pKa = 2.99LHH469 pKa = 6.29AHH471 pKa = 5.84CTRR474 pKa = 11.84EE475 pKa = 4.18LYY477 pKa = 10.68VSLMLLYY484 pKa = 8.84QTYY487 pKa = 9.1GWKK490 pKa = 10.04LHH492 pKa = 6.48LYY494 pKa = 8.45SHH496 pKa = 7.36PIVLGFRR503 pKa = 11.84KK504 pKa = 9.99VPMGLGLSPFLLAQFTSAICSVVRR528 pKa = 11.84RR529 pKa = 11.84AFPHH533 pKa = 6.38CMAFSYY539 pKa = 10.53MDD541 pKa = 4.5DD542 pKa = 3.65LVLGAKK548 pKa = 9.36SVHH551 pKa = 6.13HH552 pKa = 6.53LEE554 pKa = 4.43SLFSAITSFLLSLGIHH570 pKa = 6.68LNPEE574 pKa = 3.65KK575 pKa = 10.04TKK577 pKa = 10.52RR578 pKa = 11.84WGKK581 pKa = 9.39SLQFMGYY588 pKa = 10.21VIGAWGSLPQDD599 pKa = 4.11HH600 pKa = 6.82IRR602 pKa = 11.84AKK604 pKa = 9.95IQRR607 pKa = 11.84CFRR610 pKa = 11.84TLPCNRR616 pKa = 11.84PIDD619 pKa = 3.55WKK621 pKa = 8.96VCQRR625 pKa = 11.84IVGLLGFAAPFTQCGYY641 pKa = 8.88PALMPIYY648 pKa = 10.04HH649 pKa = 7.4AIKK652 pKa = 10.19LKK654 pKa = 10.31QAFSFSLTYY663 pKa = 10.44KK664 pKa = 10.77AFLRR668 pKa = 11.84DD669 pKa = 3.4QYY671 pKa = 11.04LTLHH675 pKa = 6.18PVARR679 pKa = 11.84QRR681 pKa = 11.84PGPCQVFADD690 pKa = 4.34ATPTGWGLVCGQQRR704 pKa = 11.84MRR706 pKa = 11.84GDD708 pKa = 3.89FVSPLPIHH716 pKa = 5.41TAEE719 pKa = 4.49LLAACFARR727 pKa = 11.84CWTGAKK733 pKa = 9.96HH734 pKa = 6.11IATDD738 pKa = 3.49NSVVLSRR745 pKa = 11.84KK746 pKa = 7.43YY747 pKa = 9.41TSFPWLLGCAANWILRR763 pKa = 11.84GTCFVYY769 pKa = 10.73VPSQQNPADD778 pKa = 4.03DD779 pKa = 4.24PSRR782 pKa = 11.84GRR784 pKa = 11.84LGLLRR789 pKa = 11.84PLPRR793 pKa = 11.84LHH795 pKa = 6.93FLPSTGRR802 pKa = 11.84TSLYY806 pKa = 10.51ADD808 pKa = 3.69SPPVPFHH815 pKa = 6.63RR816 pKa = 11.84PARR819 pKa = 11.84VHH821 pKa = 6.17FASPLHH827 pKa = 6.31DD828 pKa = 3.21AWRR831 pKa = 11.84PPP833 pKa = 3.23
Molecular weight: 94.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2097 |
153 |
833 |
349.5 |
39.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.056 ± 0.665 | 3.958 ± 0.754 |
2.957 ± 0.324 | 2.48 ± 0.54 |
5.818 ± 0.576 | 5.866 ± 0.235 |
2.909 ± 1.158 | 3.958 ± 0.692 |
2.48 ± 0.644 | 13.114 ± 0.607 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.621 ± 0.261 | 2.575 ± 0.229 |
9.68 ± 0.977 | 3.624 ± 0.341 |
5.675 ± 1.08 | 9.537 ± 0.443 |
7.058 ± 1.059 | 5.007 ± 0.404 |
3.195 ± 0.592 | 2.432 ± 0.393 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
