
Wobbly possum disease virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Arnidovirineae; Arteriviridae; Zealarterivirinae; Kappaarterivirus; Kappaarterivirus wobum
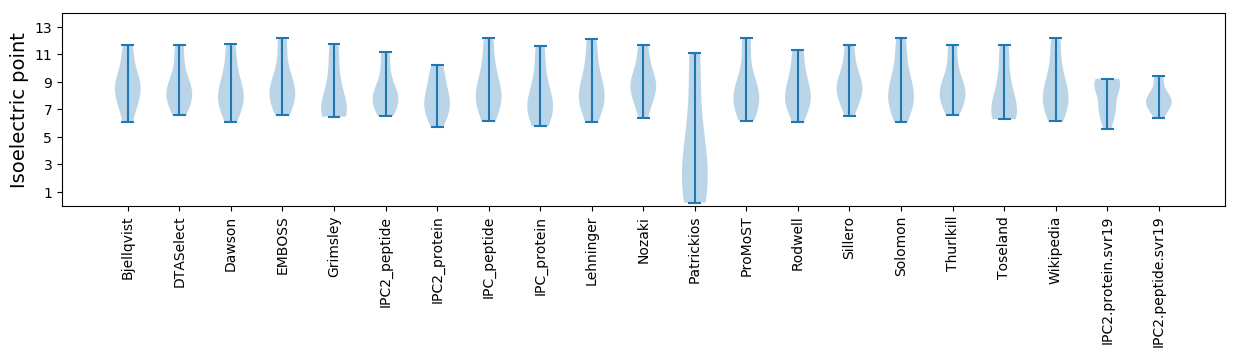
Average proteome isoelectric point is 7.94
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
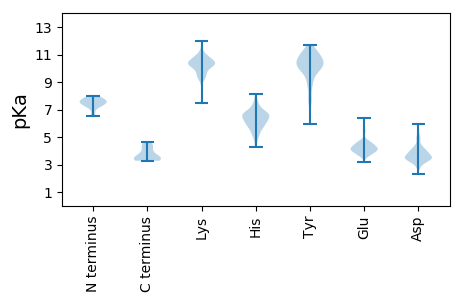
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G9FGR9|G9FGR9_9NIDO Envelope protein OS=Wobbly possum disease virus OX=1118369 PE=4 SV=1
MM1 pKa = 7.34LNWRR5 pKa = 11.84RR6 pKa = 11.84FATALCTLYY15 pKa = 11.16AFLLVRR21 pKa = 11.84TAADD25 pKa = 3.87TPDD28 pKa = 3.48PSYY31 pKa = 11.17QYY33 pKa = 10.12VHH35 pKa = 6.25AHH37 pKa = 6.38ISSCAKK43 pKa = 10.07IFQQLDD49 pKa = 2.76IDD51 pKa = 4.93FNQSLPVMLADD62 pKa = 5.98LIEE65 pKa = 4.54QPSHH69 pKa = 6.21WLSTHH74 pKa = 6.31PGVVRR79 pKa = 11.84AFDD82 pKa = 4.98EE83 pKa = 4.0IHH85 pKa = 6.61DD86 pKa = 4.34TYY88 pKa = 11.51TLATSYY94 pKa = 11.5YY95 pKa = 9.07EE96 pKa = 4.6LPHH99 pKa = 8.12SYY101 pKa = 9.76ILHH104 pKa = 5.97EE105 pKa = 4.06MLGNAEE111 pKa = 4.51AVLEE115 pKa = 3.98PLRR118 pKa = 11.84YY119 pKa = 9.05YY120 pKa = 9.18RR121 pKa = 11.84THH123 pKa = 6.24FRR125 pKa = 11.84YY126 pKa = 9.79LDD128 pKa = 3.4RR129 pKa = 11.84MALEE133 pKa = 4.47SLAHH137 pKa = 6.26LSYY140 pKa = 11.03IKK142 pKa = 10.43VQQDD146 pKa = 2.93NDD148 pKa = 3.66TLANATFTIHH158 pKa = 7.52PNIQWNEE165 pKa = 3.68LQFAGVVPSPFYY177 pKa = 9.98MVYY180 pKa = 10.37MSMACWLITIHH191 pKa = 6.95AFLRR195 pKa = 11.84LRR197 pKa = 11.84FQWILCATSRR207 pKa = 4.2
MM1 pKa = 7.34LNWRR5 pKa = 11.84RR6 pKa = 11.84FATALCTLYY15 pKa = 11.16AFLLVRR21 pKa = 11.84TAADD25 pKa = 3.87TPDD28 pKa = 3.48PSYY31 pKa = 11.17QYY33 pKa = 10.12VHH35 pKa = 6.25AHH37 pKa = 6.38ISSCAKK43 pKa = 10.07IFQQLDD49 pKa = 2.76IDD51 pKa = 4.93FNQSLPVMLADD62 pKa = 5.98LIEE65 pKa = 4.54QPSHH69 pKa = 6.21WLSTHH74 pKa = 6.31PGVVRR79 pKa = 11.84AFDD82 pKa = 4.98EE83 pKa = 4.0IHH85 pKa = 6.61DD86 pKa = 4.34TYY88 pKa = 11.51TLATSYY94 pKa = 11.5YY95 pKa = 9.07EE96 pKa = 4.6LPHH99 pKa = 8.12SYY101 pKa = 9.76ILHH104 pKa = 5.97EE105 pKa = 4.06MLGNAEE111 pKa = 4.51AVLEE115 pKa = 3.98PLRR118 pKa = 11.84YY119 pKa = 9.05YY120 pKa = 9.18RR121 pKa = 11.84THH123 pKa = 6.24FRR125 pKa = 11.84YY126 pKa = 9.79LDD128 pKa = 3.4RR129 pKa = 11.84MALEE133 pKa = 4.47SLAHH137 pKa = 6.26LSYY140 pKa = 11.03IKK142 pKa = 10.43VQQDD146 pKa = 2.93NDD148 pKa = 3.66TLANATFTIHH158 pKa = 7.52PNIQWNEE165 pKa = 3.68LQFAGVVPSPFYY177 pKa = 9.98MVYY180 pKa = 10.37MSMACWLITIHH191 pKa = 6.95AFLRR195 pKa = 11.84LRR197 pKa = 11.84FQWILCATSRR207 pKa = 4.2
Molecular weight: 24.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G9FGS4|G9FGS4_9NIDO Membrane protein OS=Wobbly possum disease virus OX=1118369 PE=4 SV=1
MM1 pKa = 6.56QTSPWSLDD9 pKa = 3.14TKK11 pKa = 11.26LMLGRR16 pKa = 11.84KK17 pKa = 7.45MVLMLKK23 pKa = 10.36LLRR26 pKa = 11.84SSSHH30 pKa = 6.56RR31 pKa = 11.84LHH33 pKa = 7.1PCHH36 pKa = 6.44TSHH39 pKa = 7.83WDD41 pKa = 3.31LTPSRR46 pKa = 11.84GCRR49 pKa = 11.84CCIIITRR56 pKa = 11.84LTIASACAYY65 pKa = 6.77TSPRR69 pKa = 11.84IFATRR74 pKa = 11.84RR75 pKa = 11.84SLRR78 pKa = 3.5
MM1 pKa = 6.56QTSPWSLDD9 pKa = 3.14TKK11 pKa = 11.26LMLGRR16 pKa = 11.84KK17 pKa = 7.45MVLMLKK23 pKa = 10.36LLRR26 pKa = 11.84SSSHH30 pKa = 6.56RR31 pKa = 11.84LHH33 pKa = 7.1PCHH36 pKa = 6.44TSHH39 pKa = 7.83WDD41 pKa = 3.31LTPSRR46 pKa = 11.84GCRR49 pKa = 11.84CCIIITRR56 pKa = 11.84LTIASACAYY65 pKa = 6.77TSPRR69 pKa = 11.84IFATRR74 pKa = 11.84RR75 pKa = 11.84SLRR78 pKa = 3.5
Molecular weight: 8.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6514 |
45 |
3402 |
651.4 |
71.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.612 ± 0.437 | 2.871 ± 0.264 |
4.421 ± 0.513 | 3.485 ± 0.387 |
4.206 ± 0.161 | 6.954 ± 0.575 |
2.702 ± 0.738 | 4.774 ± 0.346 |
3.915 ± 0.474 | 9.18 ± 0.819 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.303 ± 0.251 | 3.669 ± 0.286 |
6.739 ± 0.798 | 2.379 ± 0.203 |
4.943 ± 0.61 | 7.169 ± 0.316 |
7.016 ± 0.617 | 9.426 ± 1.054 |
1.274 ± 0.181 | 3.961 ± 0.412 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
