
Giant house spider associated circular virus 2
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.11
Get precalculated fractions of proteins
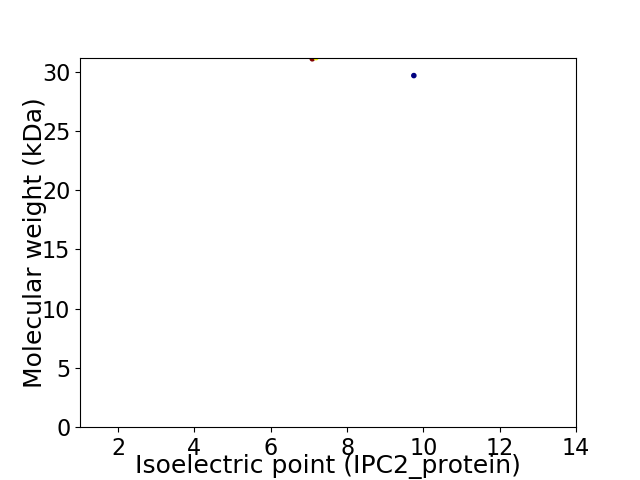
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
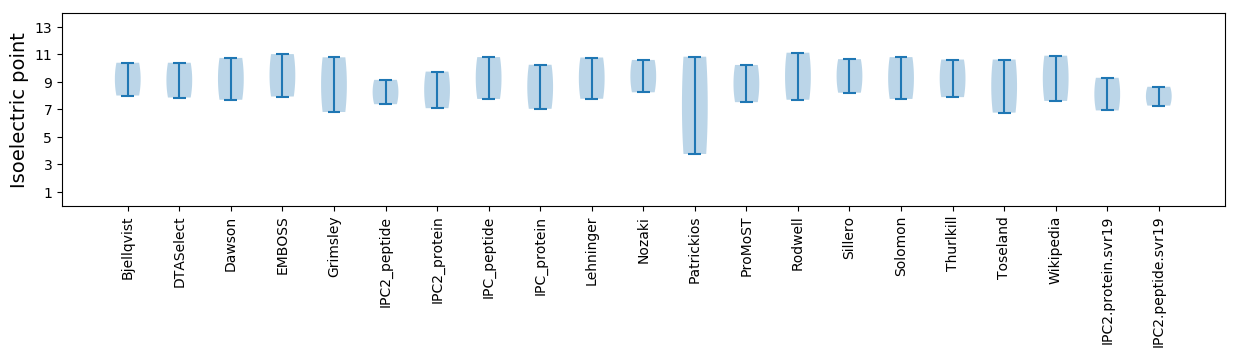
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BPD7|A0A346BPD7_9VIRU Putative capsid protein OS=Giant house spider associated circular virus 2 OX=2293289 PE=4 SV=1
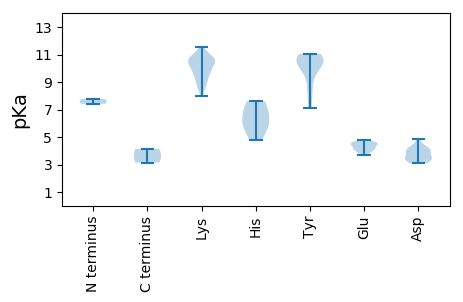
MM1 pKa = 7.42PTPKK5 pKa = 10.02KK6 pKa = 10.43AYY8 pKa = 10.23CFTLNNYY15 pKa = 7.76TSEE18 pKa = 4.46EE19 pKa = 4.12YY20 pKa = 10.9EE21 pKa = 4.36EE22 pKa = 4.46ICGKK26 pKa = 10.71LSDD29 pKa = 4.23DD30 pKa = 4.03CLYY33 pKa = 11.06AIIGKK38 pKa = 8.81EE39 pKa = 4.02VGEE42 pKa = 4.5SGTPHH47 pKa = 5.98LQGYY51 pKa = 8.65CLFKK55 pKa = 10.66RR56 pKa = 11.84ALQFNTIKK64 pKa = 10.54NRR66 pKa = 11.84YY67 pKa = 8.44LPRR70 pKa = 11.84CHH72 pKa = 7.6IEE74 pKa = 4.05VAAGSADD81 pKa = 3.43ANRR84 pKa = 11.84AYY86 pKa = 10.35CSKK89 pKa = 10.11GGSFIEE95 pKa = 4.76YY96 pKa = 7.13GTKK99 pKa = 9.76PKK101 pKa = 10.51KK102 pKa = 10.59SNTRR106 pKa = 11.84SEE108 pKa = 4.12LASAFCTHH116 pKa = 6.56MGSGRR121 pKa = 11.84SGMVQFAAEE130 pKa = 4.61EE131 pKa = 4.18PGTWYY136 pKa = 10.7FSGHH140 pKa = 5.45NLLRR144 pKa = 11.84NYY146 pKa = 10.41LAIQPPISRR155 pKa = 11.84DD156 pKa = 3.49AISVKK161 pKa = 9.53WYY163 pKa = 9.57YY164 pKa = 11.06GEE166 pKa = 4.41PGVGKK171 pKa = 9.62SRR173 pKa = 11.84RR174 pKa = 11.84AHH176 pKa = 5.98MEE178 pKa = 3.81LPDD181 pKa = 4.02AFIKK185 pKa = 10.43EE186 pKa = 4.54PRR188 pKa = 11.84TKK190 pKa = 8.81WWSGYY195 pKa = 10.37ALEE198 pKa = 4.67TDD200 pKa = 4.43VIIDD204 pKa = 3.64DD205 pKa = 4.85FGPNGIDD212 pKa = 3.32INHH215 pKa = 7.46LLRR218 pKa = 11.84WFDD221 pKa = 3.35RR222 pKa = 11.84YY223 pKa = 10.95KK224 pKa = 10.88CVVEE228 pKa = 4.57SKK230 pKa = 11.15GGMLPLYY237 pKa = 9.77AVNFIVTSNFHH248 pKa = 6.81PNQIFQFGGEE258 pKa = 4.39LNAQLPALMRR268 pKa = 11.84RR269 pKa = 11.84IEE271 pKa = 4.64CIEE274 pKa = 3.8MM275 pKa = 4.13
MM1 pKa = 7.42PTPKK5 pKa = 10.02KK6 pKa = 10.43AYY8 pKa = 10.23CFTLNNYY15 pKa = 7.76TSEE18 pKa = 4.46EE19 pKa = 4.12YY20 pKa = 10.9EE21 pKa = 4.36EE22 pKa = 4.46ICGKK26 pKa = 10.71LSDD29 pKa = 4.23DD30 pKa = 4.03CLYY33 pKa = 11.06AIIGKK38 pKa = 8.81EE39 pKa = 4.02VGEE42 pKa = 4.5SGTPHH47 pKa = 5.98LQGYY51 pKa = 8.65CLFKK55 pKa = 10.66RR56 pKa = 11.84ALQFNTIKK64 pKa = 10.54NRR66 pKa = 11.84YY67 pKa = 8.44LPRR70 pKa = 11.84CHH72 pKa = 7.6IEE74 pKa = 4.05VAAGSADD81 pKa = 3.43ANRR84 pKa = 11.84AYY86 pKa = 10.35CSKK89 pKa = 10.11GGSFIEE95 pKa = 4.76YY96 pKa = 7.13GTKK99 pKa = 9.76PKK101 pKa = 10.51KK102 pKa = 10.59SNTRR106 pKa = 11.84SEE108 pKa = 4.12LASAFCTHH116 pKa = 6.56MGSGRR121 pKa = 11.84SGMVQFAAEE130 pKa = 4.61EE131 pKa = 4.18PGTWYY136 pKa = 10.7FSGHH140 pKa = 5.45NLLRR144 pKa = 11.84NYY146 pKa = 10.41LAIQPPISRR155 pKa = 11.84DD156 pKa = 3.49AISVKK161 pKa = 9.53WYY163 pKa = 9.57YY164 pKa = 11.06GEE166 pKa = 4.41PGVGKK171 pKa = 9.62SRR173 pKa = 11.84RR174 pKa = 11.84AHH176 pKa = 5.98MEE178 pKa = 3.81LPDD181 pKa = 4.02AFIKK185 pKa = 10.43EE186 pKa = 4.54PRR188 pKa = 11.84TKK190 pKa = 8.81WWSGYY195 pKa = 10.37ALEE198 pKa = 4.67TDD200 pKa = 4.43VIIDD204 pKa = 3.64DD205 pKa = 4.85FGPNGIDD212 pKa = 3.32INHH215 pKa = 7.46LLRR218 pKa = 11.84WFDD221 pKa = 3.35RR222 pKa = 11.84YY223 pKa = 10.95KK224 pKa = 10.88CVVEE228 pKa = 4.57SKK230 pKa = 11.15GGMLPLYY237 pKa = 9.77AVNFIVTSNFHH248 pKa = 6.81PNQIFQFGGEE258 pKa = 4.39LNAQLPALMRR268 pKa = 11.84RR269 pKa = 11.84IEE271 pKa = 4.64CIEE274 pKa = 3.8MM275 pKa = 4.13
Molecular weight: 31.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPD7|A0A346BPD7_9VIRU Putative capsid protein OS=Giant house spider associated circular virus 2 OX=2293289 PE=4 SV=1
MM1 pKa = 7.76AGRR4 pKa = 11.84KK5 pKa = 8.54RR6 pKa = 11.84VYY8 pKa = 10.89AMPVSKK14 pKa = 10.66SNRR17 pKa = 11.84PFKK20 pKa = 10.38KK21 pKa = 9.77RR22 pKa = 11.84RR23 pKa = 11.84ISRR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 8.83RR29 pKa = 11.84RR30 pKa = 11.84GSRR33 pKa = 11.84KK34 pKa = 9.32SVNFTSQSGRR44 pKa = 11.84GTGLGFSRR52 pKa = 11.84KK53 pKa = 7.95RR54 pKa = 11.84TSLRR58 pKa = 11.84KK59 pKa = 9.66FKK61 pKa = 10.82RR62 pKa = 11.84LLWDD66 pKa = 3.11SSTAQTHH73 pKa = 4.82YY74 pKa = 10.51RR75 pKa = 11.84SNFAFTTAINTTAVASTMSVTVIASRR101 pKa = 11.84RR102 pKa = 11.84FNNQNFWTTAGGAINPDD119 pKa = 3.49GGTIPTFVTNGDD131 pKa = 3.37FTIRR135 pKa = 11.84GGMYY139 pKa = 10.7GLRR142 pKa = 11.84LCNAPDD148 pKa = 4.27AADD151 pKa = 3.66TDD153 pKa = 4.02KK154 pKa = 11.57DD155 pKa = 3.99AIQCIVYY162 pKa = 9.93LVKK165 pKa = 9.97STKK168 pKa = 9.42NWNSTNLPASVPVGFDD184 pKa = 3.15PSLVQDD190 pKa = 4.13FQTNIGKK197 pKa = 9.03IVYY200 pKa = 9.63KK201 pKa = 10.69KK202 pKa = 10.67EE203 pKa = 3.67FLLEE207 pKa = 4.09DD208 pKa = 3.62TNVAVIEE215 pKa = 4.11KK216 pKa = 10.51RR217 pKa = 11.84MGLSKK222 pKa = 10.11IDD224 pKa = 3.08ISEE227 pKa = 4.25YY228 pKa = 10.58QNSQSEE234 pKa = 4.58FVWIILTGIVSGTTIKK250 pKa = 10.83GVTVVPYY257 pKa = 10.61YY258 pKa = 10.74NLSFVGDD265 pKa = 3.96TVV267 pKa = 3.14
MM1 pKa = 7.76AGRR4 pKa = 11.84KK5 pKa = 8.54RR6 pKa = 11.84VYY8 pKa = 10.89AMPVSKK14 pKa = 10.66SNRR17 pKa = 11.84PFKK20 pKa = 10.38KK21 pKa = 9.77RR22 pKa = 11.84RR23 pKa = 11.84ISRR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 8.83RR29 pKa = 11.84RR30 pKa = 11.84GSRR33 pKa = 11.84KK34 pKa = 9.32SVNFTSQSGRR44 pKa = 11.84GTGLGFSRR52 pKa = 11.84KK53 pKa = 7.95RR54 pKa = 11.84TSLRR58 pKa = 11.84KK59 pKa = 9.66FKK61 pKa = 10.82RR62 pKa = 11.84LLWDD66 pKa = 3.11SSTAQTHH73 pKa = 4.82YY74 pKa = 10.51RR75 pKa = 11.84SNFAFTTAINTTAVASTMSVTVIASRR101 pKa = 11.84RR102 pKa = 11.84FNNQNFWTTAGGAINPDD119 pKa = 3.49GGTIPTFVTNGDD131 pKa = 3.37FTIRR135 pKa = 11.84GGMYY139 pKa = 10.7GLRR142 pKa = 11.84LCNAPDD148 pKa = 4.27AADD151 pKa = 3.66TDD153 pKa = 4.02KK154 pKa = 11.57DD155 pKa = 3.99AIQCIVYY162 pKa = 9.93LVKK165 pKa = 9.97STKK168 pKa = 9.42NWNSTNLPASVPVGFDD184 pKa = 3.15PSLVQDD190 pKa = 4.13FQTNIGKK197 pKa = 9.03IVYY200 pKa = 9.63KK201 pKa = 10.69KK202 pKa = 10.67EE203 pKa = 3.67FLLEE207 pKa = 4.09DD208 pKa = 3.62TNVAVIEE215 pKa = 4.11KK216 pKa = 10.51RR217 pKa = 11.84MGLSKK222 pKa = 10.11IDD224 pKa = 3.08ISEE227 pKa = 4.25YY228 pKa = 10.58QNSQSEE234 pKa = 4.58FVWIILTGIVSGTTIKK250 pKa = 10.83GVTVVPYY257 pKa = 10.61YY258 pKa = 10.74NLSFVGDD265 pKa = 3.96TVV267 pKa = 3.14
Molecular weight: 29.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
542 |
267 |
275 |
271.0 |
30.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.642 ± 0.452 | 2.03 ± 0.892 |
4.059 ± 0.303 | 4.428 ± 1.78 |
5.351 ± 0.186 | 8.303 ± 0.305 |
1.476 ± 0.767 | 6.458 ± 0.063 |
6.273 ± 0.326 | 6.273 ± 0.717 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.214 ± 0.238 | 5.72 ± 0.451 |
4.428 ± 0.736 | 2.768 ± 0.159 |
6.642 ± 0.852 | 7.934 ± 0.996 |
7.196 ± 2.031 | 5.904 ± 1.626 |
1.661 ± 0.113 | 4.244 ± 0.869 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
