
Oscillibacter sp. 1-3
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Oscillibacter; unclassified Oscillibacter
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
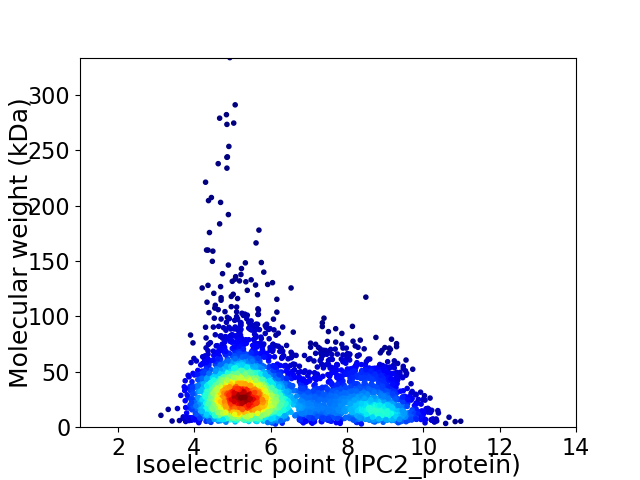
Virtual 2D-PAGE plot for 4209 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
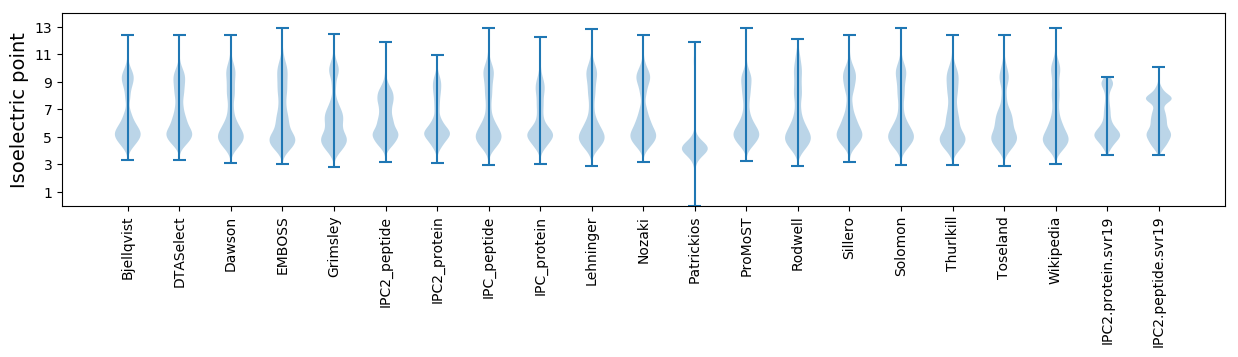
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9M9E2|R9M9E2_9FIRM N-acetyltransferase domain-containing protein OS=Oscillibacter sp. 1-3 OX=1235797 GN=C816_00196 PE=4 SV=1
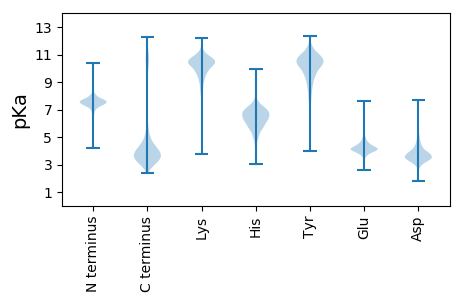
MM1 pKa = 7.06MKK3 pKa = 10.34KK4 pKa = 10.48FFGILLALAMVLSLAACGGNGGSGTPANDD33 pKa = 3.6TPPASTDD40 pKa = 3.13NNQPSGDD47 pKa = 3.87APSGGSASGTFKK59 pKa = 11.07LGGVGPLTGDD69 pKa = 3.21AAIYY73 pKa = 10.57GNAAMNGAQIAVDD86 pKa = 4.64EE87 pKa = 4.45INALGGIQIEE97 pKa = 4.99FKK99 pKa = 11.22AEE101 pKa = 3.76DD102 pKa = 3.86DD103 pKa = 4.06VADD106 pKa = 4.51GEE108 pKa = 4.82TSVNAYY114 pKa = 6.94NTLMDD119 pKa = 3.01WGMQVLVGPVTTGAAIAVSAEE140 pKa = 4.09VYY142 pKa = 10.06NDD144 pKa = 3.21RR145 pKa = 11.84VFALTPSASSTDD157 pKa = 3.78VISGKK162 pKa = 10.72DD163 pKa = 3.3NMFQVCFTDD172 pKa = 3.87PNQGVGSADD181 pKa = 3.72YY182 pKa = 10.36ISQNMPGAKK191 pKa = 9.8VGIIYY196 pKa = 10.56RR197 pKa = 11.84NDD199 pKa = 3.26DD200 pKa = 3.86AYY202 pKa = 11.56SQGIRR207 pKa = 11.84DD208 pKa = 3.89TFVAEE213 pKa = 4.0AGKK216 pKa = 10.65VGLEE220 pKa = 3.96IVSEE224 pKa = 4.51GTFTKK229 pKa = 9.81DD230 pKa = 2.99TQSDD234 pKa = 3.74FSVQLTAAQSSGADD248 pKa = 3.82LIFLPTYY255 pKa = 7.17YY256 pKa = 10.3QPNAVILNQAKK267 pKa = 10.12GMGYY271 pKa = 10.26APTFFGVDD279 pKa = 3.25GMDD282 pKa = 5.46GILAMPGFDD291 pKa = 3.04TSLAEE296 pKa = 4.23GVMLLTPFSADD307 pKa = 3.16ADD309 pKa = 4.02DD310 pKa = 4.74QRR312 pKa = 11.84TQSFVAKK319 pKa = 10.04YY320 pKa = 10.38NEE322 pKa = 4.18LHH324 pKa = 6.85GEE326 pKa = 4.22TPNQFAADD334 pKa = 4.42GYY336 pKa = 9.1DD337 pKa = 2.97AVYY340 pKa = 10.19IIYY343 pKa = 9.42EE344 pKa = 4.11ALQAAGCSGDD354 pKa = 3.32MSAEE358 pKa = 4.28EE359 pKa = 3.74ICEE362 pKa = 4.34AIVAVMPTITVDD374 pKa = 3.18GLTGQGMTWAEE385 pKa = 3.94SGEE388 pKa = 4.24VSKK391 pKa = 11.52APMAVVIKK399 pKa = 10.44DD400 pKa = 3.6GVYY403 pKa = 8.55VTPP406 pKa = 4.86
MM1 pKa = 7.06MKK3 pKa = 10.34KK4 pKa = 10.48FFGILLALAMVLSLAACGGNGGSGTPANDD33 pKa = 3.6TPPASTDD40 pKa = 3.13NNQPSGDD47 pKa = 3.87APSGGSASGTFKK59 pKa = 11.07LGGVGPLTGDD69 pKa = 3.21AAIYY73 pKa = 10.57GNAAMNGAQIAVDD86 pKa = 4.64EE87 pKa = 4.45INALGGIQIEE97 pKa = 4.99FKK99 pKa = 11.22AEE101 pKa = 3.76DD102 pKa = 3.86DD103 pKa = 4.06VADD106 pKa = 4.51GEE108 pKa = 4.82TSVNAYY114 pKa = 6.94NTLMDD119 pKa = 3.01WGMQVLVGPVTTGAAIAVSAEE140 pKa = 4.09VYY142 pKa = 10.06NDD144 pKa = 3.21RR145 pKa = 11.84VFALTPSASSTDD157 pKa = 3.78VISGKK162 pKa = 10.72DD163 pKa = 3.3NMFQVCFTDD172 pKa = 3.87PNQGVGSADD181 pKa = 3.72YY182 pKa = 10.36ISQNMPGAKK191 pKa = 9.8VGIIYY196 pKa = 10.56RR197 pKa = 11.84NDD199 pKa = 3.26DD200 pKa = 3.86AYY202 pKa = 11.56SQGIRR207 pKa = 11.84DD208 pKa = 3.89TFVAEE213 pKa = 4.0AGKK216 pKa = 10.65VGLEE220 pKa = 3.96IVSEE224 pKa = 4.51GTFTKK229 pKa = 9.81DD230 pKa = 2.99TQSDD234 pKa = 3.74FSVQLTAAQSSGADD248 pKa = 3.82LIFLPTYY255 pKa = 7.17YY256 pKa = 10.3QPNAVILNQAKK267 pKa = 10.12GMGYY271 pKa = 10.26APTFFGVDD279 pKa = 3.25GMDD282 pKa = 5.46GILAMPGFDD291 pKa = 3.04TSLAEE296 pKa = 4.23GVMLLTPFSADD307 pKa = 3.16ADD309 pKa = 4.02DD310 pKa = 4.74QRR312 pKa = 11.84TQSFVAKK319 pKa = 10.04YY320 pKa = 10.38NEE322 pKa = 4.18LHH324 pKa = 6.85GEE326 pKa = 4.22TPNQFAADD334 pKa = 4.42GYY336 pKa = 9.1DD337 pKa = 2.97AVYY340 pKa = 10.19IIYY343 pKa = 9.42EE344 pKa = 4.11ALQAAGCSGDD354 pKa = 3.32MSAEE358 pKa = 4.28EE359 pKa = 3.74ICEE362 pKa = 4.34AIVAVMPTITVDD374 pKa = 3.18GLTGQGMTWAEE385 pKa = 3.94SGEE388 pKa = 4.24VSKK391 pKa = 11.52APMAVVIKK399 pKa = 10.44DD400 pKa = 3.6GVYY403 pKa = 8.55VTPP406 pKa = 4.86
Molecular weight: 41.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9M200|R9M200_9FIRM Uncharacterized protein OS=Oscillibacter sp. 1-3 OX=1235797 GN=C816_02573 PE=4 SV=1
MM1 pKa = 7.34PRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84IQLNAVQTLALGFAALIVAGGVLLALPVSSRR36 pKa = 11.84DD37 pKa = 3.26GHH39 pKa = 6.61ALPFLDD45 pKa = 5.03ALFTAASASCVTGLVLYY62 pKa = 7.43DD63 pKa = 3.32TCTQFTLFGQAVILLLIQVGGMGFMTVSILVAVLLHH99 pKa = 6.03RR100 pKa = 11.84RR101 pKa = 11.84IGLRR105 pKa = 11.84QRR107 pKa = 11.84SILMDD112 pKa = 3.58SVGALQMGGIVRR124 pKa = 11.84LTRR127 pKa = 11.84RR128 pKa = 11.84AIRR131 pKa = 11.84VTLTCEE137 pKa = 3.84GAGAALLALWFCPRR151 pKa = 11.84YY152 pKa = 9.92GVGRR156 pKa = 11.84GLWMSLFHH164 pKa = 6.87AVSSFCNAGFDD175 pKa = 4.52LLGTGASLTSIAGEE189 pKa = 3.83PLPNIVLMALVICGGLGFLVWDD211 pKa = 4.72DD212 pKa = 4.28LLTHH216 pKa = 6.55GRR218 pKa = 11.84HH219 pKa = 4.29IRR221 pKa = 11.84RR222 pKa = 11.84WRR224 pKa = 11.84LHH226 pKa = 5.15SKK228 pKa = 9.71IVVFATTLLFVGGAAAFYY246 pKa = 10.4FLEE249 pKa = 5.07RR250 pKa = 11.84DD251 pKa = 3.37HH252 pKa = 7.41AFAGAGPGRR261 pKa = 11.84RR262 pKa = 11.84ALMAAFQSVTCRR274 pKa = 11.84TAGFNTAPLTALSQSGTLLTMILMFIGAGSGSTGGGAKK312 pKa = 10.31VNTVSVLLLSALAQVRR328 pKa = 11.84RR329 pKa = 11.84KK330 pKa = 10.17EE331 pKa = 3.97DD332 pKa = 3.12VNVFRR337 pKa = 11.84RR338 pKa = 11.84RR339 pKa = 11.84LDD341 pKa = 3.05TATIQKK347 pKa = 9.93AYY349 pKa = 10.16SSVSIFFLACLAGTMVMCLQGISLDD374 pKa = 3.7SALFEE379 pKa = 5.36AISAVGTVGLTRR391 pKa = 11.84GVTPYY396 pKa = 10.81LPEE399 pKa = 4.31LSKK402 pKa = 10.69LTVLLMMFAGRR413 pKa = 11.84VGSMSVAMAVTRR425 pKa = 11.84DD426 pKa = 3.36RR427 pKa = 11.84PQPKK431 pKa = 9.68LRR433 pKa = 11.84NIPEE437 pKa = 4.73KK438 pKa = 10.7ILIGG442 pKa = 3.81
MM1 pKa = 7.34PRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84IQLNAVQTLALGFAALIVAGGVLLALPVSSRR36 pKa = 11.84DD37 pKa = 3.26GHH39 pKa = 6.61ALPFLDD45 pKa = 5.03ALFTAASASCVTGLVLYY62 pKa = 7.43DD63 pKa = 3.32TCTQFTLFGQAVILLLIQVGGMGFMTVSILVAVLLHH99 pKa = 6.03RR100 pKa = 11.84RR101 pKa = 11.84IGLRR105 pKa = 11.84QRR107 pKa = 11.84SILMDD112 pKa = 3.58SVGALQMGGIVRR124 pKa = 11.84LTRR127 pKa = 11.84RR128 pKa = 11.84AIRR131 pKa = 11.84VTLTCEE137 pKa = 3.84GAGAALLALWFCPRR151 pKa = 11.84YY152 pKa = 9.92GVGRR156 pKa = 11.84GLWMSLFHH164 pKa = 6.87AVSSFCNAGFDD175 pKa = 4.52LLGTGASLTSIAGEE189 pKa = 3.83PLPNIVLMALVICGGLGFLVWDD211 pKa = 4.72DD212 pKa = 4.28LLTHH216 pKa = 6.55GRR218 pKa = 11.84HH219 pKa = 4.29IRR221 pKa = 11.84RR222 pKa = 11.84WRR224 pKa = 11.84LHH226 pKa = 5.15SKK228 pKa = 9.71IVVFATTLLFVGGAAAFYY246 pKa = 10.4FLEE249 pKa = 5.07RR250 pKa = 11.84DD251 pKa = 3.37HH252 pKa = 7.41AFAGAGPGRR261 pKa = 11.84RR262 pKa = 11.84ALMAAFQSVTCRR274 pKa = 11.84TAGFNTAPLTALSQSGTLLTMILMFIGAGSGSTGGGAKK312 pKa = 10.31VNTVSVLLLSALAQVRR328 pKa = 11.84RR329 pKa = 11.84KK330 pKa = 10.17EE331 pKa = 3.97DD332 pKa = 3.12VNVFRR337 pKa = 11.84RR338 pKa = 11.84RR339 pKa = 11.84LDD341 pKa = 3.05TATIQKK347 pKa = 9.93AYY349 pKa = 10.16SSVSIFFLACLAGTMVMCLQGISLDD374 pKa = 3.7SALFEE379 pKa = 5.36AISAVGTVGLTRR391 pKa = 11.84GVTPYY396 pKa = 10.81LPEE399 pKa = 4.31LSKK402 pKa = 10.69LTVLLMMFAGRR413 pKa = 11.84VGSMSVAMAVTRR425 pKa = 11.84DD426 pKa = 3.36RR427 pKa = 11.84PQPKK431 pKa = 9.68LRR433 pKa = 11.84NIPEE437 pKa = 4.73KK438 pKa = 10.7ILIGG442 pKa = 3.81
Molecular weight: 46.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1246555 |
25 |
2967 |
296.2 |
32.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.764 ± 0.05 | 1.675 ± 0.019 |
5.457 ± 0.03 | 7.055 ± 0.038 |
3.762 ± 0.029 | 7.914 ± 0.039 |
1.738 ± 0.018 | 5.497 ± 0.036 |
4.563 ± 0.038 | 9.875 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.704 ± 0.02 | 3.285 ± 0.024 |
4.303 ± 0.031 | 3.511 ± 0.026 |
6.192 ± 0.048 | 5.577 ± 0.025 |
5.61 ± 0.05 | 6.86 ± 0.035 |
1.138 ± 0.016 | 3.518 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
