
Humulus japonicus latent virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Ilarvirus
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
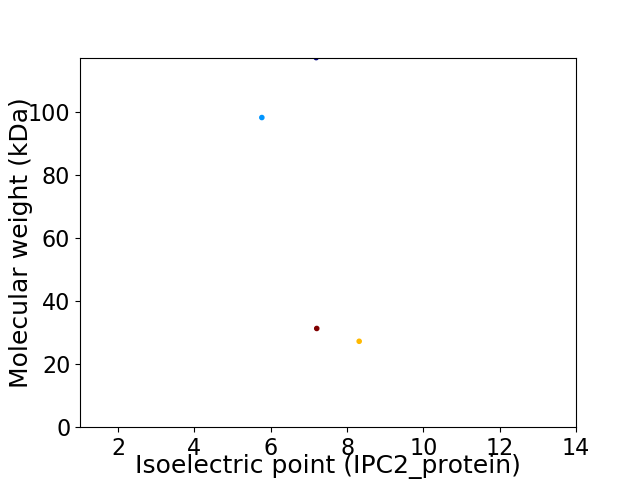
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q6JE42|Q6JE42_9BROM ATP-dependent helicase OS=Humulus japonicus latent virus OX=269213 PE=3 SV=1
MM1 pKa = 7.78ASFKK5 pKa = 11.0DD6 pKa = 3.25IVNALFASAQQKK18 pKa = 10.53LGLSVPLLTGDD29 pKa = 4.09KK30 pKa = 10.14IIVNMPSNQKK40 pKa = 10.72KK41 pKa = 10.2KK42 pKa = 9.17MEE44 pKa = 4.25LVAVPLLRR52 pKa = 11.84EE53 pKa = 3.75NVRR56 pKa = 11.84FSRR59 pKa = 11.84EE60 pKa = 3.8TLVVFLRR67 pKa = 11.84HH68 pKa = 5.85PCLANMLNQWEE79 pKa = 4.58FWAWEE84 pKa = 4.17FNEE87 pKa = 3.87EE88 pKa = 4.22LLPYY92 pKa = 8.73GCSVTRR98 pKa = 11.84VKK100 pKa = 10.42DD101 pKa = 3.17ICVDD105 pKa = 3.62GPYY108 pKa = 9.96EE109 pKa = 4.29DD110 pKa = 3.75KK111 pKa = 10.97SGYY114 pKa = 9.4FHH116 pKa = 7.74GSYY119 pKa = 9.77TYY121 pKa = 11.14AEE123 pKa = 4.04NSVYY127 pKa = 10.61VDD129 pKa = 3.94DD130 pKa = 4.9YY131 pKa = 11.52QVEE134 pKa = 4.65LCPLDD139 pKa = 5.5DD140 pKa = 4.19NMPPEE145 pKa = 4.19SVYY148 pKa = 11.11EE149 pKa = 3.99EE150 pKa = 5.15GEE152 pKa = 4.3CSSKK156 pKa = 10.39YY157 pKa = 9.95QEE159 pKa = 4.38VPPCEE164 pKa = 4.37DD165 pKa = 3.13VNLDD169 pKa = 3.6GEE171 pKa = 4.27EE172 pKa = 4.45GEE174 pKa = 4.62NVGLLDD180 pKa = 3.84MPEE183 pKa = 4.77KK184 pKa = 10.74DD185 pKa = 4.16FQSLMHH191 pKa = 6.49TRR193 pKa = 11.84LQHH196 pKa = 6.23LRR198 pKa = 11.84DD199 pKa = 3.77VGCSINLSGRR209 pKa = 11.84GIPVIPIVAGGFSEE223 pKa = 4.39TTRR226 pKa = 11.84LNPRR230 pKa = 11.84HH231 pKa = 6.15AEE233 pKa = 3.9SEE235 pKa = 4.34RR236 pKa = 11.84VFKK239 pKa = 10.92DD240 pKa = 3.01RR241 pKa = 11.84LEE243 pKa = 4.32GPEE246 pKa = 4.61LKK248 pKa = 9.83WAGPVFKK255 pKa = 10.72DD256 pKa = 3.12NLRR259 pKa = 11.84AHH261 pKa = 6.63SVDD264 pKa = 4.03DD265 pKa = 3.39MDD267 pKa = 3.47VRR269 pKa = 11.84EE270 pKa = 4.22WVAINRR276 pKa = 11.84PIPKK280 pKa = 9.42KK281 pKa = 9.81KK282 pKa = 10.55LPMKK286 pKa = 10.41ISGSIDD292 pKa = 3.31APLLNVRR299 pKa = 11.84EE300 pKa = 4.11VSPVLNLVNNYY311 pKa = 8.83IPKK314 pKa = 9.56YY315 pKa = 10.9QMLDD319 pKa = 2.58WHH321 pKa = 6.75ALAKK325 pKa = 10.23FINNYY330 pKa = 6.6QAKK333 pKa = 8.76EE334 pKa = 3.86KK335 pKa = 10.78PMDD338 pKa = 3.25IKK340 pKa = 10.12ITSRR344 pKa = 11.84GFANRR349 pKa = 11.84HH350 pKa = 4.6VFQQALDD357 pKa = 4.29EE358 pKa = 4.65ILPHH362 pKa = 5.72HH363 pKa = 6.83HH364 pKa = 6.89KK365 pKa = 10.66FDD367 pKa = 4.29DD368 pKa = 4.01SFFQTMVEE376 pKa = 4.29CDD378 pKa = 4.32DD379 pKa = 3.95ISIEE383 pKa = 4.59LSRR386 pKa = 11.84CRR388 pKa = 11.84LDD390 pKa = 4.45LSVFNSWEE398 pKa = 3.9KK399 pKa = 11.01GSNKK403 pKa = 8.76VKK405 pKa = 10.5PMINSGVANRR415 pKa = 11.84RR416 pKa = 11.84FSTQRR421 pKa = 11.84EE422 pKa = 3.93AMLAVQKK429 pKa = 11.12RR430 pKa = 11.84NMNVPNLAHH439 pKa = 6.36CTDD442 pKa = 3.45IPAVVAEE449 pKa = 4.36VVDD452 pKa = 4.1KK453 pKa = 10.29FFKK456 pKa = 11.01VIIDD460 pKa = 3.85PDD462 pKa = 3.91KK463 pKa = 11.12LSQFPGFISEE473 pKa = 4.48GEE475 pKa = 3.76LAYY478 pKa = 10.88FEE480 pKa = 5.83DD481 pKa = 4.68YY482 pKa = 11.27LRR484 pKa = 11.84GKK486 pKa = 10.12QPDD489 pKa = 3.89PEE491 pKa = 4.2MLKK494 pKa = 10.86DD495 pKa = 3.54PLCFQRR501 pKa = 11.84LDD503 pKa = 3.57KK504 pKa = 10.38YY505 pKa = 11.2RR506 pKa = 11.84HH507 pKa = 4.36MMKK510 pKa = 10.86NNMKK514 pKa = 9.92PVEE517 pKa = 4.07DD518 pKa = 4.01HH519 pKa = 6.52SLNSEE524 pKa = 4.06RR525 pKa = 11.84ALMSTITYY533 pKa = 9.29HH534 pKa = 6.17EE535 pKa = 5.21KK536 pKa = 9.08GTIMSTSPIFLMAANRR552 pKa = 11.84ILMCLSDD559 pKa = 3.95KK560 pKa = 11.27VKK562 pKa = 10.6IPTGKK567 pKa = 9.18YY568 pKa = 8.52HH569 pKa = 7.04QLFSLDD575 pKa = 3.27AHH577 pKa = 5.63VLKK580 pKa = 10.61HH581 pKa = 4.96VKK583 pKa = 8.67HH584 pKa = 5.87WKK586 pKa = 10.21EE587 pKa = 3.22IDD589 pKa = 3.37FSKK592 pKa = 10.59FDD594 pKa = 3.37KK595 pKa = 11.19SQGEE599 pKa = 4.05LHH601 pKa = 6.94HH602 pKa = 5.92YY603 pKa = 7.97LQRR606 pKa = 11.84EE607 pKa = 3.69IFIRR611 pKa = 11.84IGVPMNFIDD620 pKa = 3.32AWVPQVTRR628 pKa = 11.84SFISDD633 pKa = 3.77PSNGLFFNVDD643 pKa = 3.8FQRR646 pKa = 11.84RR647 pKa = 11.84TGDD650 pKa = 2.74ACTYY654 pKa = 10.46LGNTLVTLCVLCHH667 pKa = 6.3VYY669 pKa = 10.92DD670 pKa = 4.55LGDD673 pKa = 3.96ANVLSVVASGDD684 pKa = 3.36DD685 pKa = 3.74CLIGSYY691 pKa = 10.52EE692 pKa = 4.25KK693 pKa = 10.71LDD695 pKa = 3.36QSRR698 pKa = 11.84EE699 pKa = 4.13YY700 pKa = 11.15LCSTLFNFEE709 pKa = 4.4AKK711 pKa = 10.25FPYY714 pKa = 9.38NQPFICSKK722 pKa = 10.29FLIIAEE728 pKa = 4.1MMDD731 pKa = 3.01GRR733 pKa = 11.84EE734 pKa = 3.94EE735 pKa = 4.29VIAVPNPIKK744 pKa = 10.72LLIKK748 pKa = 10.4LGSKK752 pKa = 10.36LNPEE756 pKa = 5.23DD757 pKa = 3.61FDD759 pKa = 3.66EE760 pKa = 5.38WFLAWRR766 pKa = 11.84DD767 pKa = 3.8HH768 pKa = 5.65IHH770 pKa = 6.03YY771 pKa = 8.43FQNAHH776 pKa = 6.54IVDD779 pKa = 4.14QIVDD783 pKa = 4.01LVEE786 pKa = 3.87VRR788 pKa = 11.84TGVRR792 pKa = 11.84NTQCLRR798 pKa = 11.84LALEE802 pKa = 4.32SFPHH806 pKa = 5.64VFRR809 pKa = 11.84SKK811 pKa = 9.9KK812 pKa = 9.7AVMRR816 pKa = 11.84EE817 pKa = 3.97FFCMEE822 pKa = 4.44EE823 pKa = 3.86IEE825 pKa = 4.47SIEE828 pKa = 4.04RR829 pKa = 11.84NRR831 pKa = 11.84QIKK834 pKa = 9.61AIKK837 pKa = 9.98ALNKK841 pKa = 9.86KK842 pKa = 9.9FKK844 pKa = 10.46LPRR847 pKa = 11.84STTHH851 pKa = 4.86VCC853 pKa = 3.58
MM1 pKa = 7.78ASFKK5 pKa = 11.0DD6 pKa = 3.25IVNALFASAQQKK18 pKa = 10.53LGLSVPLLTGDD29 pKa = 4.09KK30 pKa = 10.14IIVNMPSNQKK40 pKa = 10.72KK41 pKa = 10.2KK42 pKa = 9.17MEE44 pKa = 4.25LVAVPLLRR52 pKa = 11.84EE53 pKa = 3.75NVRR56 pKa = 11.84FSRR59 pKa = 11.84EE60 pKa = 3.8TLVVFLRR67 pKa = 11.84HH68 pKa = 5.85PCLANMLNQWEE79 pKa = 4.58FWAWEE84 pKa = 4.17FNEE87 pKa = 3.87EE88 pKa = 4.22LLPYY92 pKa = 8.73GCSVTRR98 pKa = 11.84VKK100 pKa = 10.42DD101 pKa = 3.17ICVDD105 pKa = 3.62GPYY108 pKa = 9.96EE109 pKa = 4.29DD110 pKa = 3.75KK111 pKa = 10.97SGYY114 pKa = 9.4FHH116 pKa = 7.74GSYY119 pKa = 9.77TYY121 pKa = 11.14AEE123 pKa = 4.04NSVYY127 pKa = 10.61VDD129 pKa = 3.94DD130 pKa = 4.9YY131 pKa = 11.52QVEE134 pKa = 4.65LCPLDD139 pKa = 5.5DD140 pKa = 4.19NMPPEE145 pKa = 4.19SVYY148 pKa = 11.11EE149 pKa = 3.99EE150 pKa = 5.15GEE152 pKa = 4.3CSSKK156 pKa = 10.39YY157 pKa = 9.95QEE159 pKa = 4.38VPPCEE164 pKa = 4.37DD165 pKa = 3.13VNLDD169 pKa = 3.6GEE171 pKa = 4.27EE172 pKa = 4.45GEE174 pKa = 4.62NVGLLDD180 pKa = 3.84MPEE183 pKa = 4.77KK184 pKa = 10.74DD185 pKa = 4.16FQSLMHH191 pKa = 6.49TRR193 pKa = 11.84LQHH196 pKa = 6.23LRR198 pKa = 11.84DD199 pKa = 3.77VGCSINLSGRR209 pKa = 11.84GIPVIPIVAGGFSEE223 pKa = 4.39TTRR226 pKa = 11.84LNPRR230 pKa = 11.84HH231 pKa = 6.15AEE233 pKa = 3.9SEE235 pKa = 4.34RR236 pKa = 11.84VFKK239 pKa = 10.92DD240 pKa = 3.01RR241 pKa = 11.84LEE243 pKa = 4.32GPEE246 pKa = 4.61LKK248 pKa = 9.83WAGPVFKK255 pKa = 10.72DD256 pKa = 3.12NLRR259 pKa = 11.84AHH261 pKa = 6.63SVDD264 pKa = 4.03DD265 pKa = 3.39MDD267 pKa = 3.47VRR269 pKa = 11.84EE270 pKa = 4.22WVAINRR276 pKa = 11.84PIPKK280 pKa = 9.42KK281 pKa = 9.81KK282 pKa = 10.55LPMKK286 pKa = 10.41ISGSIDD292 pKa = 3.31APLLNVRR299 pKa = 11.84EE300 pKa = 4.11VSPVLNLVNNYY311 pKa = 8.83IPKK314 pKa = 9.56YY315 pKa = 10.9QMLDD319 pKa = 2.58WHH321 pKa = 6.75ALAKK325 pKa = 10.23FINNYY330 pKa = 6.6QAKK333 pKa = 8.76EE334 pKa = 3.86KK335 pKa = 10.78PMDD338 pKa = 3.25IKK340 pKa = 10.12ITSRR344 pKa = 11.84GFANRR349 pKa = 11.84HH350 pKa = 4.6VFQQALDD357 pKa = 4.29EE358 pKa = 4.65ILPHH362 pKa = 5.72HH363 pKa = 6.83HH364 pKa = 6.89KK365 pKa = 10.66FDD367 pKa = 4.29DD368 pKa = 4.01SFFQTMVEE376 pKa = 4.29CDD378 pKa = 4.32DD379 pKa = 3.95ISIEE383 pKa = 4.59LSRR386 pKa = 11.84CRR388 pKa = 11.84LDD390 pKa = 4.45LSVFNSWEE398 pKa = 3.9KK399 pKa = 11.01GSNKK403 pKa = 8.76VKK405 pKa = 10.5PMINSGVANRR415 pKa = 11.84RR416 pKa = 11.84FSTQRR421 pKa = 11.84EE422 pKa = 3.93AMLAVQKK429 pKa = 11.12RR430 pKa = 11.84NMNVPNLAHH439 pKa = 6.36CTDD442 pKa = 3.45IPAVVAEE449 pKa = 4.36VVDD452 pKa = 4.1KK453 pKa = 10.29FFKK456 pKa = 11.01VIIDD460 pKa = 3.85PDD462 pKa = 3.91KK463 pKa = 11.12LSQFPGFISEE473 pKa = 4.48GEE475 pKa = 3.76LAYY478 pKa = 10.88FEE480 pKa = 5.83DD481 pKa = 4.68YY482 pKa = 11.27LRR484 pKa = 11.84GKK486 pKa = 10.12QPDD489 pKa = 3.89PEE491 pKa = 4.2MLKK494 pKa = 10.86DD495 pKa = 3.54PLCFQRR501 pKa = 11.84LDD503 pKa = 3.57KK504 pKa = 10.38YY505 pKa = 11.2RR506 pKa = 11.84HH507 pKa = 4.36MMKK510 pKa = 10.86NNMKK514 pKa = 9.92PVEE517 pKa = 4.07DD518 pKa = 4.01HH519 pKa = 6.52SLNSEE524 pKa = 4.06RR525 pKa = 11.84ALMSTITYY533 pKa = 9.29HH534 pKa = 6.17EE535 pKa = 5.21KK536 pKa = 9.08GTIMSTSPIFLMAANRR552 pKa = 11.84ILMCLSDD559 pKa = 3.95KK560 pKa = 11.27VKK562 pKa = 10.6IPTGKK567 pKa = 9.18YY568 pKa = 8.52HH569 pKa = 7.04QLFSLDD575 pKa = 3.27AHH577 pKa = 5.63VLKK580 pKa = 10.61HH581 pKa = 4.96VKK583 pKa = 8.67HH584 pKa = 5.87WKK586 pKa = 10.21EE587 pKa = 3.22IDD589 pKa = 3.37FSKK592 pKa = 10.59FDD594 pKa = 3.37KK595 pKa = 11.19SQGEE599 pKa = 4.05LHH601 pKa = 6.94HH602 pKa = 5.92YY603 pKa = 7.97LQRR606 pKa = 11.84EE607 pKa = 3.69IFIRR611 pKa = 11.84IGVPMNFIDD620 pKa = 3.32AWVPQVTRR628 pKa = 11.84SFISDD633 pKa = 3.77PSNGLFFNVDD643 pKa = 3.8FQRR646 pKa = 11.84RR647 pKa = 11.84TGDD650 pKa = 2.74ACTYY654 pKa = 10.46LGNTLVTLCVLCHH667 pKa = 6.3VYY669 pKa = 10.92DD670 pKa = 4.55LGDD673 pKa = 3.96ANVLSVVASGDD684 pKa = 3.36DD685 pKa = 3.74CLIGSYY691 pKa = 10.52EE692 pKa = 4.25KK693 pKa = 10.71LDD695 pKa = 3.36QSRR698 pKa = 11.84EE699 pKa = 4.13YY700 pKa = 11.15LCSTLFNFEE709 pKa = 4.4AKK711 pKa = 10.25FPYY714 pKa = 9.38NQPFICSKK722 pKa = 10.29FLIIAEE728 pKa = 4.1MMDD731 pKa = 3.01GRR733 pKa = 11.84EE734 pKa = 3.94EE735 pKa = 4.29VIAVPNPIKK744 pKa = 10.72LLIKK748 pKa = 10.4LGSKK752 pKa = 10.36LNPEE756 pKa = 5.23DD757 pKa = 3.61FDD759 pKa = 3.66EE760 pKa = 5.38WFLAWRR766 pKa = 11.84DD767 pKa = 3.8HH768 pKa = 5.65IHH770 pKa = 6.03YY771 pKa = 8.43FQNAHH776 pKa = 6.54IVDD779 pKa = 4.14QIVDD783 pKa = 4.01LVEE786 pKa = 3.87VRR788 pKa = 11.84TGVRR792 pKa = 11.84NTQCLRR798 pKa = 11.84LALEE802 pKa = 4.32SFPHH806 pKa = 5.64VFRR809 pKa = 11.84SKK811 pKa = 9.9KK812 pKa = 9.7AVMRR816 pKa = 11.84EE817 pKa = 3.97FFCMEE822 pKa = 4.44EE823 pKa = 3.86IEE825 pKa = 4.47SIEE828 pKa = 4.04RR829 pKa = 11.84NRR831 pKa = 11.84QIKK834 pKa = 9.61AIKK837 pKa = 9.98ALNKK841 pKa = 9.86KK842 pKa = 9.9FKK844 pKa = 10.46LPRR847 pKa = 11.84STTHH851 pKa = 4.86VCC853 pKa = 3.58
Molecular weight: 98.14 kDa
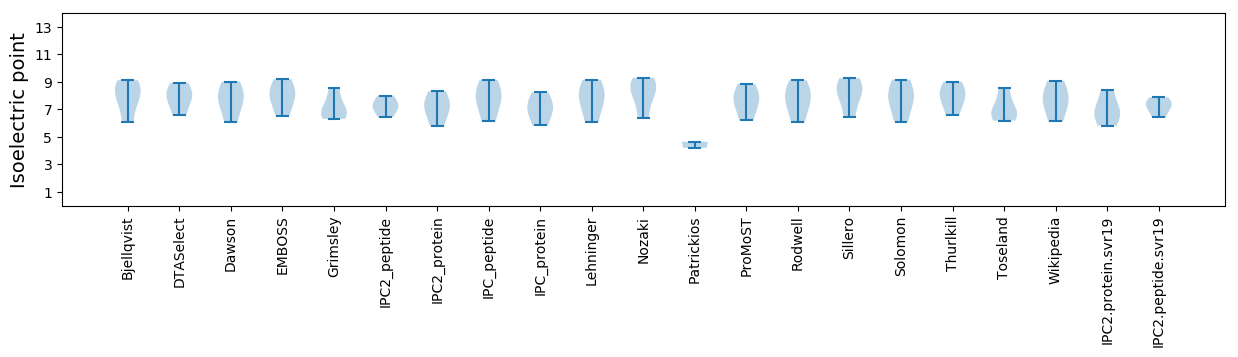
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6JE40|Q6JE40_9BROM Movement protein OS=Humulus japonicus latent virus OX=269213 PE=4 SV=1
MM1 pKa = 7.54SNAFCASCGLRR12 pKa = 11.84SHH14 pKa = 7.36VGNCPPRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 9.97ASPGQKK29 pKa = 9.65NRR31 pKa = 11.84TRR33 pKa = 11.84IYY35 pKa = 9.49ALEE38 pKa = 4.06RR39 pKa = 11.84KK40 pKa = 8.8RR41 pKa = 11.84ANVRR45 pKa = 11.84VPPGTTLNPKK55 pKa = 9.88FFAQFSGYY63 pKa = 9.12NAEE66 pKa = 4.31EE67 pKa = 4.66CDD69 pKa = 5.09LLDD72 pKa = 5.05DD73 pKa = 5.03VINAIPSVSVSAASQKK89 pKa = 10.47RR90 pKa = 11.84VTEE93 pKa = 4.13AQLVVGDD100 pKa = 4.45PLKK103 pKa = 10.88GIQGPCTHH111 pKa = 6.78MGDD114 pKa = 3.43ATITARR120 pKa = 11.84TGGVRR125 pKa = 11.84GFYY128 pKa = 9.26MALDD132 pKa = 3.99ALLTGIPDD140 pKa = 3.5ATRR143 pKa = 11.84LKK145 pKa = 10.43SLVMHH150 pKa = 6.33ITFLSDD156 pKa = 3.39GGVLGLNTEE165 pKa = 4.23RR166 pKa = 11.84SDD168 pKa = 4.22QYY170 pKa = 11.77VEE172 pKa = 4.09PNPLKK177 pKa = 10.47RR178 pKa = 11.84KK179 pKa = 9.39RR180 pKa = 11.84FEE182 pKa = 4.03KK183 pKa = 10.49EE184 pKa = 3.8SPFWAQILFPDD195 pKa = 3.9NVKK198 pKa = 10.83LSDD201 pKa = 3.54VKK203 pKa = 11.17NYY205 pKa = 11.12AIMIKK210 pKa = 10.22FDD212 pKa = 3.64SDD214 pKa = 3.71YY215 pKa = 11.47SANTPLYY222 pKa = 10.16TRR224 pKa = 11.84EE225 pKa = 3.97CWVNHH230 pKa = 5.13YY231 pKa = 8.88QLPSAVIPEE240 pKa = 3.81EE241 pKa = 3.88AFARR245 pKa = 11.84KK246 pKa = 9.06
MM1 pKa = 7.54SNAFCASCGLRR12 pKa = 11.84SHH14 pKa = 7.36VGNCPPRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 9.97ASPGQKK29 pKa = 9.65NRR31 pKa = 11.84TRR33 pKa = 11.84IYY35 pKa = 9.49ALEE38 pKa = 4.06RR39 pKa = 11.84KK40 pKa = 8.8RR41 pKa = 11.84ANVRR45 pKa = 11.84VPPGTTLNPKK55 pKa = 9.88FFAQFSGYY63 pKa = 9.12NAEE66 pKa = 4.31EE67 pKa = 4.66CDD69 pKa = 5.09LLDD72 pKa = 5.05DD73 pKa = 5.03VINAIPSVSVSAASQKK89 pKa = 10.47RR90 pKa = 11.84VTEE93 pKa = 4.13AQLVVGDD100 pKa = 4.45PLKK103 pKa = 10.88GIQGPCTHH111 pKa = 6.78MGDD114 pKa = 3.43ATITARR120 pKa = 11.84TGGVRR125 pKa = 11.84GFYY128 pKa = 9.26MALDD132 pKa = 3.99ALLTGIPDD140 pKa = 3.5ATRR143 pKa = 11.84LKK145 pKa = 10.43SLVMHH150 pKa = 6.33ITFLSDD156 pKa = 3.39GGVLGLNTEE165 pKa = 4.23RR166 pKa = 11.84SDD168 pKa = 4.22QYY170 pKa = 11.77VEE172 pKa = 4.09PNPLKK177 pKa = 10.47RR178 pKa = 11.84KK179 pKa = 9.39RR180 pKa = 11.84FEE182 pKa = 4.03KK183 pKa = 10.49EE184 pKa = 3.8SPFWAQILFPDD195 pKa = 3.9NVKK198 pKa = 10.83LSDD201 pKa = 3.54VKK203 pKa = 11.17NYY205 pKa = 11.12AIMIKK210 pKa = 10.22FDD212 pKa = 3.64SDD214 pKa = 3.71YY215 pKa = 11.47SANTPLYY222 pKa = 10.16TRR224 pKa = 11.84EE225 pKa = 3.97CWVNHH230 pKa = 5.13YY231 pKa = 8.88QLPSAVIPEE240 pKa = 3.81EE241 pKa = 3.88AFARR245 pKa = 11.84KK246 pKa = 9.06
Molecular weight: 27.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2418 |
246 |
1041 |
604.5 |
68.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.692 ± 1.174 | 2.605 ± 0.148 |
6.245 ± 0.292 | 6.452 ± 0.416 |
4.839 ± 0.412 | 4.384 ± 0.362 |
2.481 ± 0.301 | 5.873 ± 0.252 |
7.155 ± 0.429 | 8.768 ± 0.26 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.771 ± 0.295 | 4.715 ± 0.331 |
4.715 ± 0.605 | 2.978 ± 0.249 |
5.624 ± 0.307 | 6.865 ± 0.259 |
4.549 ± 0.631 | 7.279 ± 0.194 |
1.034 ± 0.113 | 2.978 ± 0.18 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
