
Amasya cherry disease associated chrysovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Chrysoviridae; Alphachrysovirus
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
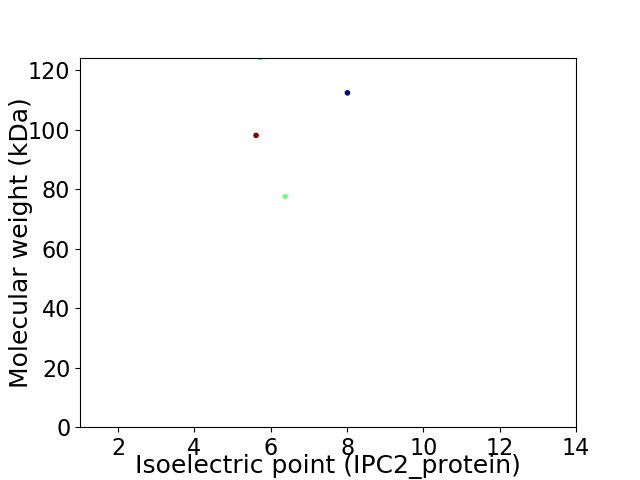
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
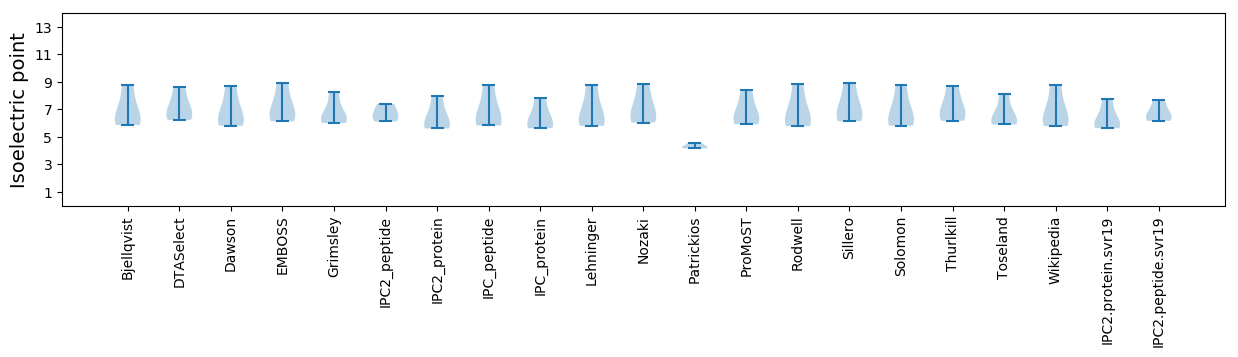
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q65A80|Q65A80_9VIRU Uncharacterized protein OS=Amasya cherry disease associated chrysovirus OX=284687 PE=4 SV=1
MM1 pKa = 7.39SAFTSVFDD9 pKa = 4.76HH10 pKa = 6.49MVTASNKK17 pKa = 9.79LNHH20 pKa = 6.0EE21 pKa = 4.33ALWRR25 pKa = 11.84ASLKK29 pKa = 11.21GEE31 pKa = 4.11GAVQEE36 pKa = 4.2DD37 pKa = 4.11TRR39 pKa = 11.84DD40 pKa = 3.55LFVEE44 pKa = 5.02EE45 pKa = 4.87ISVISQTEE53 pKa = 4.03KK54 pKa = 11.07NVLSGCDD61 pKa = 3.27MPAKK65 pKa = 10.7YY66 pKa = 9.46EE67 pKa = 4.06IKK69 pKa = 10.69GMAEE73 pKa = 3.71NGLKK77 pKa = 10.81LEE79 pKa = 4.42VTAVAHH85 pKa = 6.53GDD87 pKa = 3.46AVGKK91 pKa = 10.34FDD93 pKa = 3.48ITHH96 pKa = 6.45SKK98 pKa = 8.52TEE100 pKa = 3.84KK101 pKa = 9.22LTYY104 pKa = 10.14ASEE107 pKa = 4.52VEE109 pKa = 3.97WALRR113 pKa = 11.84PRR115 pKa = 11.84GCRR118 pKa = 11.84LNYY121 pKa = 9.57PRR123 pKa = 11.84ANDD126 pKa = 3.63SMLSRR131 pKa = 11.84NKK133 pKa = 10.06RR134 pKa = 11.84VLGEE138 pKa = 3.92LQAACVNLSEE148 pKa = 5.59SEE150 pKa = 4.07QSDD153 pKa = 3.97LGITLAAEE161 pKa = 4.3KK162 pKa = 10.33LYY164 pKa = 9.79STSDD168 pKa = 3.09MRR170 pKa = 11.84HH171 pKa = 6.48ILTKK175 pKa = 9.71MYY177 pKa = 11.23LMLCDD182 pKa = 4.01LTLADD187 pKa = 4.39TSASTRR193 pKa = 11.84WLYY196 pKa = 11.73AMGASKK202 pKa = 11.14FEE204 pKa = 4.12MPKK207 pKa = 9.42LTGGQLIGAIVDD219 pKa = 3.5YY220 pKa = 11.2DD221 pKa = 4.08VVVDD225 pKa = 4.42KK226 pKa = 11.53AGFSNEE232 pKa = 3.96EE233 pKa = 4.04LTMLWLGAMEE243 pKa = 4.16YY244 pKa = 10.13PSVKK248 pKa = 9.8FCRR251 pKa = 11.84DD252 pKa = 2.87NVYY255 pKa = 9.08NTIRR259 pKa = 11.84MEE261 pKa = 4.56ADD263 pKa = 2.95SVAVVSKK270 pKa = 10.74NQFVLEE276 pKa = 4.02RR277 pKa = 11.84HH278 pKa = 5.78MNWDD282 pKa = 3.53PEE284 pKa = 4.27TMYY287 pKa = 10.82RR288 pKa = 11.84AIAGICCKK296 pKa = 10.09LDD298 pKa = 4.69CIGDD302 pKa = 3.48WFDD305 pKa = 3.18VVKK308 pKa = 10.97NMRR311 pKa = 11.84GLPFLMADD319 pKa = 3.43VVEE322 pKa = 4.48KK323 pKa = 10.22TGRR326 pKa = 11.84TTFNSAIPKK335 pKa = 6.82STNYY339 pKa = 9.35EE340 pKa = 3.81RR341 pKa = 11.84ALGGTALWKK350 pKa = 10.43HH351 pKa = 5.95VIRR354 pKa = 11.84RR355 pKa = 11.84NPGYY359 pKa = 9.37IATSIGIVADD369 pKa = 3.42AMMGEE374 pKa = 4.49VLKK377 pKa = 10.33MATTMVVEE385 pKa = 3.87QLGGYY390 pKa = 6.76GTLMIPKK397 pKa = 9.16KK398 pKa = 10.36APSSFEE404 pKa = 3.82YY405 pKa = 10.53DD406 pKa = 2.92SLMRR410 pKa = 11.84DD411 pKa = 3.45YY412 pKa = 11.25GLGMDD417 pKa = 4.89DD418 pKa = 4.17PSLNALLLAWSNVRR432 pKa = 11.84QQDD435 pKa = 3.14VRR437 pKa = 11.84IAFGRR442 pKa = 11.84TLIEE446 pKa = 4.09YY447 pKa = 9.42FKK449 pKa = 10.71VTTTRR454 pKa = 11.84MRR456 pKa = 11.84RR457 pKa = 11.84GQEE460 pKa = 3.43KK461 pKa = 10.52VMPQLTFDD469 pKa = 3.68MSYY472 pKa = 11.17LPIDD476 pKa = 3.52MCSWTLIKK484 pKa = 10.39GVPHH488 pKa = 7.53DD489 pKa = 4.28GFQLGMKK496 pKa = 10.19NSEE499 pKa = 3.87RR500 pKa = 11.84DD501 pKa = 3.47DD502 pKa = 3.95LEE504 pKa = 3.97RR505 pKa = 11.84ARR507 pKa = 11.84AALLWGMGVRR517 pKa = 11.84EE518 pKa = 4.1EE519 pKa = 4.32RR520 pKa = 11.84PHH522 pKa = 7.82VFANKK527 pKa = 10.35DD528 pKa = 3.06KK529 pKa = 11.16DD530 pKa = 3.94GEE532 pKa = 4.24DD533 pKa = 3.85MITSAEE539 pKa = 4.03RR540 pKa = 11.84KK541 pKa = 9.57VMSGTTGVIKK551 pKa = 10.18LSWMTYY557 pKa = 10.35ALSDD561 pKa = 3.74SFKK564 pKa = 10.28PRR566 pKa = 11.84QDD568 pKa = 3.27VAEE571 pKa = 4.83GEE573 pKa = 4.2ASSLIGVAIKK583 pKa = 8.79GTRR586 pKa = 11.84CEE588 pKa = 3.8VFYY591 pKa = 11.06KK592 pKa = 10.7GGSEE596 pKa = 3.63WDD598 pKa = 3.29VRR600 pKa = 11.84TLVEE604 pKa = 4.45PGVQAGVAQTMGFSVAPVMVKK625 pKa = 10.38EE626 pKa = 4.03PLGFDD631 pKa = 3.94GIKK634 pKa = 10.49AITAGGSAVIRR645 pKa = 11.84GKK647 pKa = 10.84KK648 pKa = 8.81PVADD652 pKa = 2.98IFKK655 pKa = 10.41NDD657 pKa = 3.33RR658 pKa = 11.84RR659 pKa = 11.84DD660 pKa = 3.58SATSTVKK667 pKa = 10.86LKK669 pKa = 10.82VSKK672 pKa = 10.88AQMKK676 pKa = 9.21KK677 pKa = 9.98PAWIDD682 pKa = 3.24YY683 pKa = 10.27DD684 pKa = 4.11GNDD687 pKa = 3.42VGEE690 pKa = 4.38KK691 pKa = 9.83KK692 pKa = 10.07HH693 pKa = 6.23YY694 pKa = 8.99CDD696 pKa = 4.17KK697 pKa = 10.88VAEE700 pKa = 4.28KK701 pKa = 10.69SEE703 pKa = 4.16KK704 pKa = 9.23VQQGDD709 pKa = 3.74TIKK712 pKa = 10.78LMRR715 pKa = 11.84VSTPGDD721 pKa = 3.78GKK723 pKa = 11.05CGLHH727 pKa = 7.62AIAQSLNLKK736 pKa = 10.15GAVKK740 pKa = 10.4QDD742 pKa = 4.07DD743 pKa = 3.82IKK745 pKa = 11.56DD746 pKa = 3.58AFDD749 pKa = 4.35NLDD752 pKa = 3.72KK753 pKa = 10.96LAQGKK758 pKa = 6.61TWHH761 pKa = 6.96EE762 pKa = 4.32DD763 pKa = 3.18NVLGALCEE771 pKa = 4.04QIGFGLRR778 pKa = 11.84LYY780 pKa = 10.44DD781 pKa = 3.62VNGEE785 pKa = 4.07NVVLHH790 pKa = 6.93RR791 pKa = 11.84YY792 pKa = 8.16EE793 pKa = 5.14KK794 pKa = 11.41DD795 pKa = 2.68MDD797 pKa = 3.93AVVDD801 pKa = 3.78VMRR804 pKa = 11.84EE805 pKa = 3.7NGHH808 pKa = 5.9YY809 pKa = 7.63EE810 pKa = 4.05TVIPNTDD817 pKa = 2.28KK818 pKa = 10.94GYY820 pKa = 10.19RR821 pKa = 11.84YY822 pKa = 9.83AVSAVFDD829 pKa = 4.56LEE831 pKa = 4.43TEE833 pKa = 4.23PEE835 pKa = 4.07RR836 pKa = 11.84MHH838 pKa = 7.1GVDD841 pKa = 5.18APITAIKK848 pKa = 10.18SITAASDD855 pKa = 3.33DD856 pKa = 3.93GKK858 pKa = 11.44GDD860 pKa = 2.99IMKK863 pKa = 10.3RR864 pKa = 11.84ATSDD868 pKa = 2.97WEE870 pKa = 4.66KK871 pKa = 10.47PQDD874 pKa = 3.45WDD876 pKa = 4.15GKK878 pKa = 9.16PGSHH882 pKa = 6.17GWW884 pKa = 3.26
MM1 pKa = 7.39SAFTSVFDD9 pKa = 4.76HH10 pKa = 6.49MVTASNKK17 pKa = 9.79LNHH20 pKa = 6.0EE21 pKa = 4.33ALWRR25 pKa = 11.84ASLKK29 pKa = 11.21GEE31 pKa = 4.11GAVQEE36 pKa = 4.2DD37 pKa = 4.11TRR39 pKa = 11.84DD40 pKa = 3.55LFVEE44 pKa = 5.02EE45 pKa = 4.87ISVISQTEE53 pKa = 4.03KK54 pKa = 11.07NVLSGCDD61 pKa = 3.27MPAKK65 pKa = 10.7YY66 pKa = 9.46EE67 pKa = 4.06IKK69 pKa = 10.69GMAEE73 pKa = 3.71NGLKK77 pKa = 10.81LEE79 pKa = 4.42VTAVAHH85 pKa = 6.53GDD87 pKa = 3.46AVGKK91 pKa = 10.34FDD93 pKa = 3.48ITHH96 pKa = 6.45SKK98 pKa = 8.52TEE100 pKa = 3.84KK101 pKa = 9.22LTYY104 pKa = 10.14ASEE107 pKa = 4.52VEE109 pKa = 3.97WALRR113 pKa = 11.84PRR115 pKa = 11.84GCRR118 pKa = 11.84LNYY121 pKa = 9.57PRR123 pKa = 11.84ANDD126 pKa = 3.63SMLSRR131 pKa = 11.84NKK133 pKa = 10.06RR134 pKa = 11.84VLGEE138 pKa = 3.92LQAACVNLSEE148 pKa = 5.59SEE150 pKa = 4.07QSDD153 pKa = 3.97LGITLAAEE161 pKa = 4.3KK162 pKa = 10.33LYY164 pKa = 9.79STSDD168 pKa = 3.09MRR170 pKa = 11.84HH171 pKa = 6.48ILTKK175 pKa = 9.71MYY177 pKa = 11.23LMLCDD182 pKa = 4.01LTLADD187 pKa = 4.39TSASTRR193 pKa = 11.84WLYY196 pKa = 11.73AMGASKK202 pKa = 11.14FEE204 pKa = 4.12MPKK207 pKa = 9.42LTGGQLIGAIVDD219 pKa = 3.5YY220 pKa = 11.2DD221 pKa = 4.08VVVDD225 pKa = 4.42KK226 pKa = 11.53AGFSNEE232 pKa = 3.96EE233 pKa = 4.04LTMLWLGAMEE243 pKa = 4.16YY244 pKa = 10.13PSVKK248 pKa = 9.8FCRR251 pKa = 11.84DD252 pKa = 2.87NVYY255 pKa = 9.08NTIRR259 pKa = 11.84MEE261 pKa = 4.56ADD263 pKa = 2.95SVAVVSKK270 pKa = 10.74NQFVLEE276 pKa = 4.02RR277 pKa = 11.84HH278 pKa = 5.78MNWDD282 pKa = 3.53PEE284 pKa = 4.27TMYY287 pKa = 10.82RR288 pKa = 11.84AIAGICCKK296 pKa = 10.09LDD298 pKa = 4.69CIGDD302 pKa = 3.48WFDD305 pKa = 3.18VVKK308 pKa = 10.97NMRR311 pKa = 11.84GLPFLMADD319 pKa = 3.43VVEE322 pKa = 4.48KK323 pKa = 10.22TGRR326 pKa = 11.84TTFNSAIPKK335 pKa = 6.82STNYY339 pKa = 9.35EE340 pKa = 3.81RR341 pKa = 11.84ALGGTALWKK350 pKa = 10.43HH351 pKa = 5.95VIRR354 pKa = 11.84RR355 pKa = 11.84NPGYY359 pKa = 9.37IATSIGIVADD369 pKa = 3.42AMMGEE374 pKa = 4.49VLKK377 pKa = 10.33MATTMVVEE385 pKa = 3.87QLGGYY390 pKa = 6.76GTLMIPKK397 pKa = 9.16KK398 pKa = 10.36APSSFEE404 pKa = 3.82YY405 pKa = 10.53DD406 pKa = 2.92SLMRR410 pKa = 11.84DD411 pKa = 3.45YY412 pKa = 11.25GLGMDD417 pKa = 4.89DD418 pKa = 4.17PSLNALLLAWSNVRR432 pKa = 11.84QQDD435 pKa = 3.14VRR437 pKa = 11.84IAFGRR442 pKa = 11.84TLIEE446 pKa = 4.09YY447 pKa = 9.42FKK449 pKa = 10.71VTTTRR454 pKa = 11.84MRR456 pKa = 11.84RR457 pKa = 11.84GQEE460 pKa = 3.43KK461 pKa = 10.52VMPQLTFDD469 pKa = 3.68MSYY472 pKa = 11.17LPIDD476 pKa = 3.52MCSWTLIKK484 pKa = 10.39GVPHH488 pKa = 7.53DD489 pKa = 4.28GFQLGMKK496 pKa = 10.19NSEE499 pKa = 3.87RR500 pKa = 11.84DD501 pKa = 3.47DD502 pKa = 3.95LEE504 pKa = 3.97RR505 pKa = 11.84ARR507 pKa = 11.84AALLWGMGVRR517 pKa = 11.84EE518 pKa = 4.1EE519 pKa = 4.32RR520 pKa = 11.84PHH522 pKa = 7.82VFANKK527 pKa = 10.35DD528 pKa = 3.06KK529 pKa = 11.16DD530 pKa = 3.94GEE532 pKa = 4.24DD533 pKa = 3.85MITSAEE539 pKa = 4.03RR540 pKa = 11.84KK541 pKa = 9.57VMSGTTGVIKK551 pKa = 10.18LSWMTYY557 pKa = 10.35ALSDD561 pKa = 3.74SFKK564 pKa = 10.28PRR566 pKa = 11.84QDD568 pKa = 3.27VAEE571 pKa = 4.83GEE573 pKa = 4.2ASSLIGVAIKK583 pKa = 8.79GTRR586 pKa = 11.84CEE588 pKa = 3.8VFYY591 pKa = 11.06KK592 pKa = 10.7GGSEE596 pKa = 3.63WDD598 pKa = 3.29VRR600 pKa = 11.84TLVEE604 pKa = 4.45PGVQAGVAQTMGFSVAPVMVKK625 pKa = 10.38EE626 pKa = 4.03PLGFDD631 pKa = 3.94GIKK634 pKa = 10.49AITAGGSAVIRR645 pKa = 11.84GKK647 pKa = 10.84KK648 pKa = 8.81PVADD652 pKa = 2.98IFKK655 pKa = 10.41NDD657 pKa = 3.33RR658 pKa = 11.84RR659 pKa = 11.84DD660 pKa = 3.58SATSTVKK667 pKa = 10.86LKK669 pKa = 10.82VSKK672 pKa = 10.88AQMKK676 pKa = 9.21KK677 pKa = 9.98PAWIDD682 pKa = 3.24YY683 pKa = 10.27DD684 pKa = 4.11GNDD687 pKa = 3.42VGEE690 pKa = 4.38KK691 pKa = 9.83KK692 pKa = 10.07HH693 pKa = 6.23YY694 pKa = 8.99CDD696 pKa = 4.17KK697 pKa = 10.88VAEE700 pKa = 4.28KK701 pKa = 10.69SEE703 pKa = 4.16KK704 pKa = 9.23VQQGDD709 pKa = 3.74TIKK712 pKa = 10.78LMRR715 pKa = 11.84VSTPGDD721 pKa = 3.78GKK723 pKa = 11.05CGLHH727 pKa = 7.62AIAQSLNLKK736 pKa = 10.15GAVKK740 pKa = 10.4QDD742 pKa = 4.07DD743 pKa = 3.82IKK745 pKa = 11.56DD746 pKa = 3.58AFDD749 pKa = 4.35NLDD752 pKa = 3.72KK753 pKa = 10.96LAQGKK758 pKa = 6.61TWHH761 pKa = 6.96EE762 pKa = 4.32DD763 pKa = 3.18NVLGALCEE771 pKa = 4.04QIGFGLRR778 pKa = 11.84LYY780 pKa = 10.44DD781 pKa = 3.62VNGEE785 pKa = 4.07NVVLHH790 pKa = 6.93RR791 pKa = 11.84YY792 pKa = 8.16EE793 pKa = 5.14KK794 pKa = 11.41DD795 pKa = 2.68MDD797 pKa = 3.93AVVDD801 pKa = 3.78VMRR804 pKa = 11.84EE805 pKa = 3.7NGHH808 pKa = 5.9YY809 pKa = 7.63EE810 pKa = 4.05TVIPNTDD817 pKa = 2.28KK818 pKa = 10.94GYY820 pKa = 10.19RR821 pKa = 11.84YY822 pKa = 9.83AVSAVFDD829 pKa = 4.56LEE831 pKa = 4.43TEE833 pKa = 4.23PEE835 pKa = 4.07RR836 pKa = 11.84MHH838 pKa = 7.1GVDD841 pKa = 5.18APITAIKK848 pKa = 10.18SITAASDD855 pKa = 3.33DD856 pKa = 3.93GKK858 pKa = 11.44GDD860 pKa = 2.99IMKK863 pKa = 10.3RR864 pKa = 11.84ATSDD868 pKa = 2.97WEE870 pKa = 4.66KK871 pKa = 10.47PQDD874 pKa = 3.45WDD876 pKa = 4.15GKK878 pKa = 9.16PGSHH882 pKa = 6.17GWW884 pKa = 3.26
Molecular weight: 98.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q65A79|Q65A79_9VIRU Putative protease OS=Amasya cherry disease associated chrysovirus OX=284687 PE=4 SV=1
MM1 pKa = 6.76TTFSRR6 pKa = 11.84EE7 pKa = 4.05QYY9 pKa = 10.25SKK11 pKa = 10.64RR12 pKa = 11.84ASLFSSLRR20 pKa = 11.84ARR22 pKa = 11.84GNATRR27 pKa = 11.84AQFAKK32 pKa = 9.97IDD34 pKa = 3.52NWKK37 pKa = 8.62EE38 pKa = 3.37WSVADD43 pKa = 3.51STAVVGQKK51 pKa = 8.68KK52 pKa = 8.39TGWTHH57 pKa = 5.63TFGEE61 pKa = 4.31AFSYY65 pKa = 11.02VDD67 pKa = 3.39RR68 pKa = 11.84CSLGGVDD75 pKa = 4.22ALSRR79 pKa = 11.84SLLRR83 pKa = 11.84ASYY86 pKa = 11.07SVFGDD91 pKa = 3.35LRR93 pKa = 11.84RR94 pKa = 11.84SVALGEE100 pKa = 4.08PTQGLEE106 pKa = 4.12YY107 pKa = 9.55TFKK110 pKa = 9.02WTRR113 pKa = 11.84ADD115 pKa = 3.34ISAEE119 pKa = 3.82LHH121 pKa = 6.02TGDD124 pKa = 4.3VCVTTPAEE132 pKa = 4.03GLMTVVSTNGRR143 pKa = 11.84DD144 pKa = 3.05RR145 pKa = 11.84SSISRR150 pKa = 11.84ATGWDD155 pKa = 3.41RR156 pKa = 11.84NSCSNMTSSDD166 pKa = 3.03VTTFINLARR175 pKa = 11.84ASCAGGNRR183 pKa = 11.84FTRR186 pKa = 11.84LVKK189 pKa = 10.7GCLIYY194 pKa = 9.83MDD196 pKa = 4.84LMYY199 pKa = 11.01GGDD202 pKa = 3.36ARR204 pKa = 11.84VSTEE208 pKa = 3.78LKK210 pKa = 10.79NLIEE214 pKa = 4.26YY215 pKa = 10.08EE216 pKa = 4.16PEE218 pKa = 4.11PVLMSYY224 pKa = 10.15RR225 pKa = 11.84VSGRR229 pKa = 11.84SYY231 pKa = 11.05AYY233 pKa = 10.14SSQNTSVEE241 pKa = 4.36HH242 pKa = 6.6ILLLSLMGNRR252 pKa = 11.84YY253 pKa = 8.73PNKK256 pKa = 9.81EE257 pKa = 3.94VPTTCRR263 pKa = 11.84ATIPEE268 pKa = 4.81DD269 pKa = 3.52GDD271 pKa = 3.73HH272 pKa = 5.76YY273 pKa = 10.94VVVRR277 pKa = 11.84GAIRR281 pKa = 11.84HH282 pKa = 5.38LSGSRR287 pKa = 11.84TVTLTYY293 pKa = 10.28RR294 pKa = 11.84GVYY297 pKa = 8.98SALVKK302 pKa = 10.43YY303 pKa = 9.75AAEE306 pKa = 4.1VGSPNDD312 pKa = 4.02LEE314 pKa = 4.29PAMVVASSMYY324 pKa = 9.89HH325 pKa = 5.14NRR327 pKa = 11.84YY328 pKa = 7.71LQKK331 pKa = 10.58VSLPSVSSVNEE342 pKa = 4.27LIMPAMVEE350 pKa = 3.65NDD352 pKa = 3.36VAVVSRR358 pKa = 11.84PHH360 pKa = 6.16VNKK363 pKa = 8.51EE364 pKa = 3.86TLATIGKK371 pKa = 7.83LHH373 pKa = 6.12QMSLLLMIKK382 pKa = 10.22DD383 pKa = 3.66VIIAAKK389 pKa = 10.14VSTRR393 pKa = 11.84SEE395 pKa = 3.54LDD397 pKa = 3.16YY398 pKa = 11.24RR399 pKa = 11.84RR400 pKa = 11.84VAMQWLNNYY409 pKa = 9.36DD410 pKa = 3.67MLMSRR415 pKa = 11.84YY416 pKa = 9.01NVNATALKK424 pKa = 10.3LVEE427 pKa = 4.92ATPQMKK433 pKa = 9.42WSYY436 pKa = 10.72LINRR440 pKa = 11.84DD441 pKa = 4.71DD442 pKa = 6.49LIDD445 pKa = 4.5LDD447 pKa = 4.76GISMFEE453 pKa = 4.24CLWLCEE459 pKa = 5.01DD460 pKa = 4.39GTLCMKK466 pKa = 10.69NGGIIMMKK474 pKa = 9.88NAKK477 pKa = 9.64KK478 pKa = 10.31DD479 pKa = 3.41GRR481 pKa = 11.84AQNPYY486 pKa = 10.94LEE488 pKa = 4.22ILKK491 pKa = 10.68DD492 pKa = 3.86EE493 pKa = 4.33IKK495 pKa = 10.22KK496 pKa = 10.65GRR498 pKa = 11.84IIYY501 pKa = 7.49DD502 pKa = 3.2TRR504 pKa = 11.84RR505 pKa = 11.84IPDD508 pKa = 3.4GDD510 pKa = 3.53FTIAGRR516 pKa = 11.84YY517 pKa = 8.15IVSDD521 pKa = 3.76EE522 pKa = 4.09DD523 pKa = 4.25AMPKK527 pKa = 9.1TIQYY531 pKa = 10.15SEE533 pKa = 3.78VRR535 pKa = 11.84IKK537 pKa = 10.59RR538 pKa = 11.84KK539 pKa = 9.92KK540 pKa = 9.82GDD542 pKa = 3.56YY543 pKa = 10.87AVVKK547 pKa = 9.84PKK549 pKa = 10.69ARR551 pKa = 11.84YY552 pKa = 9.94DD553 pKa = 3.5EE554 pKa = 5.08DD555 pKa = 4.06GCIEE559 pKa = 4.0EE560 pKa = 5.53LYY562 pKa = 10.41DD563 pKa = 4.31GSMEE567 pKa = 4.1KK568 pKa = 10.44PMSGFTEE575 pKa = 4.4TEE577 pKa = 3.59RR578 pKa = 11.84SEE580 pKa = 3.96MSEE583 pKa = 3.9EE584 pKa = 4.03YY585 pKa = 8.17MTEE588 pKa = 3.72KK589 pKa = 10.71EE590 pKa = 4.45EE591 pKa = 4.35YY592 pKa = 8.91LTAPSNVTPYY602 pKa = 11.35DD603 pKa = 3.52EE604 pKa = 5.34FKK606 pKa = 10.74LDD608 pKa = 3.31RR609 pKa = 11.84PMPVMLSRR617 pKa = 11.84PSIKK621 pKa = 9.99PRR623 pKa = 11.84FDD625 pKa = 3.12ARR627 pKa = 11.84KK628 pKa = 7.73TPFKK632 pKa = 10.1PLRR635 pKa = 11.84SSWGKK640 pKa = 10.71KK641 pKa = 9.65SDD643 pKa = 3.74DD644 pKa = 3.47EE645 pKa = 4.18PRR647 pKa = 11.84YY648 pKa = 9.39EE649 pKa = 4.64GPSKK653 pKa = 11.05SIDD656 pKa = 3.36SSPSPKK662 pKa = 10.16NKK664 pKa = 9.32NRR666 pKa = 11.84GLIVVSDD673 pKa = 3.85TEE675 pKa = 4.45GSGKK679 pKa = 10.04EE680 pKa = 3.82SAAEE684 pKa = 3.89KK685 pKa = 10.19VSPVRR690 pKa = 11.84PANRR694 pKa = 11.84RR695 pKa = 11.84SIVSVAKK702 pKa = 10.18MIEE705 pKa = 4.0DD706 pKa = 3.0KK707 pKa = 10.38MAEE710 pKa = 3.69VRR712 pKa = 11.84ARR714 pKa = 11.84EE715 pKa = 4.04KK716 pKa = 11.01ARR718 pKa = 11.84TLSPPRR724 pKa = 11.84QTGEE728 pKa = 3.97VEE730 pKa = 3.69ADD732 pKa = 3.06KK733 pKa = 10.26PQQGEE738 pKa = 3.89ADD740 pKa = 3.41TSEE743 pKa = 4.32KK744 pKa = 10.71VDD746 pKa = 3.54VPEE749 pKa = 4.3TKK751 pKa = 10.6EE752 pKa = 3.71MDD754 pKa = 4.09KK755 pKa = 10.78PALPANVAKK764 pKa = 10.16QVSVTMPEE772 pKa = 3.73KK773 pKa = 10.29STEE776 pKa = 3.03KK777 pKa = 9.46WRR779 pKa = 11.84RR780 pKa = 11.84LKK782 pKa = 10.89EE783 pKa = 3.98PTNTQIPDD791 pKa = 3.29EE792 pKa = 4.21FLVDD796 pKa = 3.89NKK798 pKa = 10.86NDD800 pKa = 3.98LKK802 pKa = 10.65WRR804 pKa = 11.84EE805 pKa = 3.76INIDD809 pKa = 3.63KK810 pKa = 10.63EE811 pKa = 4.18NEE813 pKa = 3.72EE814 pKa = 4.66MITMEE819 pKa = 4.7SRR821 pKa = 11.84VEE823 pKa = 3.95QVNEE827 pKa = 3.35ADD829 pKa = 3.51KK830 pKa = 10.8RR831 pKa = 11.84AKK833 pKa = 9.94EE834 pKa = 3.9EE835 pKa = 3.69RR836 pKa = 11.84AEE838 pKa = 4.2RR839 pKa = 11.84VPADD843 pKa = 4.07KK844 pKa = 11.05KK845 pKa = 9.88SDD847 pKa = 3.42AYY849 pKa = 11.24RR850 pKa = 11.84MLVEE854 pKa = 4.8KK855 pKa = 10.91NVDD858 pKa = 3.31PTKK861 pKa = 10.96KK862 pKa = 10.32LFGMSAAKK870 pKa = 9.25WLEE873 pKa = 3.4YY874 pKa = 10.84MMTNKK879 pKa = 10.0PKK881 pKa = 10.49GYY883 pKa = 8.63YY884 pKa = 9.73EE885 pKa = 4.43VMRR888 pKa = 11.84SDD890 pKa = 3.14INEE893 pKa = 4.66FISKK897 pKa = 8.11YY898 pKa = 8.05TATTIAPKK906 pKa = 10.29HH907 pKa = 5.89ILDD910 pKa = 4.0WVPFADD916 pKa = 3.25SVQINGALRR925 pKa = 11.84SARR928 pKa = 11.84RR929 pKa = 11.84SQTSGTNYY937 pKa = 10.15LFSEE941 pKa = 4.34SDD943 pKa = 3.43STIMNVALNIIRR955 pKa = 11.84KK956 pKa = 9.48SGGEE960 pKa = 3.81VWPVSMKK967 pKa = 10.92YY968 pKa = 8.37MGRR971 pKa = 11.84MQRR974 pKa = 11.84EE975 pKa = 4.1FYY977 pKa = 10.27VNKK980 pKa = 9.89VDD982 pKa = 3.45RR983 pKa = 11.84TAYY986 pKa = 10.2NLAAQKK992 pKa = 10.89HH993 pKa = 4.6MM994 pKa = 4.36
MM1 pKa = 6.76TTFSRR6 pKa = 11.84EE7 pKa = 4.05QYY9 pKa = 10.25SKK11 pKa = 10.64RR12 pKa = 11.84ASLFSSLRR20 pKa = 11.84ARR22 pKa = 11.84GNATRR27 pKa = 11.84AQFAKK32 pKa = 9.97IDD34 pKa = 3.52NWKK37 pKa = 8.62EE38 pKa = 3.37WSVADD43 pKa = 3.51STAVVGQKK51 pKa = 8.68KK52 pKa = 8.39TGWTHH57 pKa = 5.63TFGEE61 pKa = 4.31AFSYY65 pKa = 11.02VDD67 pKa = 3.39RR68 pKa = 11.84CSLGGVDD75 pKa = 4.22ALSRR79 pKa = 11.84SLLRR83 pKa = 11.84ASYY86 pKa = 11.07SVFGDD91 pKa = 3.35LRR93 pKa = 11.84RR94 pKa = 11.84SVALGEE100 pKa = 4.08PTQGLEE106 pKa = 4.12YY107 pKa = 9.55TFKK110 pKa = 9.02WTRR113 pKa = 11.84ADD115 pKa = 3.34ISAEE119 pKa = 3.82LHH121 pKa = 6.02TGDD124 pKa = 4.3VCVTTPAEE132 pKa = 4.03GLMTVVSTNGRR143 pKa = 11.84DD144 pKa = 3.05RR145 pKa = 11.84SSISRR150 pKa = 11.84ATGWDD155 pKa = 3.41RR156 pKa = 11.84NSCSNMTSSDD166 pKa = 3.03VTTFINLARR175 pKa = 11.84ASCAGGNRR183 pKa = 11.84FTRR186 pKa = 11.84LVKK189 pKa = 10.7GCLIYY194 pKa = 9.83MDD196 pKa = 4.84LMYY199 pKa = 11.01GGDD202 pKa = 3.36ARR204 pKa = 11.84VSTEE208 pKa = 3.78LKK210 pKa = 10.79NLIEE214 pKa = 4.26YY215 pKa = 10.08EE216 pKa = 4.16PEE218 pKa = 4.11PVLMSYY224 pKa = 10.15RR225 pKa = 11.84VSGRR229 pKa = 11.84SYY231 pKa = 11.05AYY233 pKa = 10.14SSQNTSVEE241 pKa = 4.36HH242 pKa = 6.6ILLLSLMGNRR252 pKa = 11.84YY253 pKa = 8.73PNKK256 pKa = 9.81EE257 pKa = 3.94VPTTCRR263 pKa = 11.84ATIPEE268 pKa = 4.81DD269 pKa = 3.52GDD271 pKa = 3.73HH272 pKa = 5.76YY273 pKa = 10.94VVVRR277 pKa = 11.84GAIRR281 pKa = 11.84HH282 pKa = 5.38LSGSRR287 pKa = 11.84TVTLTYY293 pKa = 10.28RR294 pKa = 11.84GVYY297 pKa = 8.98SALVKK302 pKa = 10.43YY303 pKa = 9.75AAEE306 pKa = 4.1VGSPNDD312 pKa = 4.02LEE314 pKa = 4.29PAMVVASSMYY324 pKa = 9.89HH325 pKa = 5.14NRR327 pKa = 11.84YY328 pKa = 7.71LQKK331 pKa = 10.58VSLPSVSSVNEE342 pKa = 4.27LIMPAMVEE350 pKa = 3.65NDD352 pKa = 3.36VAVVSRR358 pKa = 11.84PHH360 pKa = 6.16VNKK363 pKa = 8.51EE364 pKa = 3.86TLATIGKK371 pKa = 7.83LHH373 pKa = 6.12QMSLLLMIKK382 pKa = 10.22DD383 pKa = 3.66VIIAAKK389 pKa = 10.14VSTRR393 pKa = 11.84SEE395 pKa = 3.54LDD397 pKa = 3.16YY398 pKa = 11.24RR399 pKa = 11.84RR400 pKa = 11.84VAMQWLNNYY409 pKa = 9.36DD410 pKa = 3.67MLMSRR415 pKa = 11.84YY416 pKa = 9.01NVNATALKK424 pKa = 10.3LVEE427 pKa = 4.92ATPQMKK433 pKa = 9.42WSYY436 pKa = 10.72LINRR440 pKa = 11.84DD441 pKa = 4.71DD442 pKa = 6.49LIDD445 pKa = 4.5LDD447 pKa = 4.76GISMFEE453 pKa = 4.24CLWLCEE459 pKa = 5.01DD460 pKa = 4.39GTLCMKK466 pKa = 10.69NGGIIMMKK474 pKa = 9.88NAKK477 pKa = 9.64KK478 pKa = 10.31DD479 pKa = 3.41GRR481 pKa = 11.84AQNPYY486 pKa = 10.94LEE488 pKa = 4.22ILKK491 pKa = 10.68DD492 pKa = 3.86EE493 pKa = 4.33IKK495 pKa = 10.22KK496 pKa = 10.65GRR498 pKa = 11.84IIYY501 pKa = 7.49DD502 pKa = 3.2TRR504 pKa = 11.84RR505 pKa = 11.84IPDD508 pKa = 3.4GDD510 pKa = 3.53FTIAGRR516 pKa = 11.84YY517 pKa = 8.15IVSDD521 pKa = 3.76EE522 pKa = 4.09DD523 pKa = 4.25AMPKK527 pKa = 9.1TIQYY531 pKa = 10.15SEE533 pKa = 3.78VRR535 pKa = 11.84IKK537 pKa = 10.59RR538 pKa = 11.84KK539 pKa = 9.92KK540 pKa = 9.82GDD542 pKa = 3.56YY543 pKa = 10.87AVVKK547 pKa = 9.84PKK549 pKa = 10.69ARR551 pKa = 11.84YY552 pKa = 9.94DD553 pKa = 3.5EE554 pKa = 5.08DD555 pKa = 4.06GCIEE559 pKa = 4.0EE560 pKa = 5.53LYY562 pKa = 10.41DD563 pKa = 4.31GSMEE567 pKa = 4.1KK568 pKa = 10.44PMSGFTEE575 pKa = 4.4TEE577 pKa = 3.59RR578 pKa = 11.84SEE580 pKa = 3.96MSEE583 pKa = 3.9EE584 pKa = 4.03YY585 pKa = 8.17MTEE588 pKa = 3.72KK589 pKa = 10.71EE590 pKa = 4.45EE591 pKa = 4.35YY592 pKa = 8.91LTAPSNVTPYY602 pKa = 11.35DD603 pKa = 3.52EE604 pKa = 5.34FKK606 pKa = 10.74LDD608 pKa = 3.31RR609 pKa = 11.84PMPVMLSRR617 pKa = 11.84PSIKK621 pKa = 9.99PRR623 pKa = 11.84FDD625 pKa = 3.12ARR627 pKa = 11.84KK628 pKa = 7.73TPFKK632 pKa = 10.1PLRR635 pKa = 11.84SSWGKK640 pKa = 10.71KK641 pKa = 9.65SDD643 pKa = 3.74DD644 pKa = 3.47EE645 pKa = 4.18PRR647 pKa = 11.84YY648 pKa = 9.39EE649 pKa = 4.64GPSKK653 pKa = 11.05SIDD656 pKa = 3.36SSPSPKK662 pKa = 10.16NKK664 pKa = 9.32NRR666 pKa = 11.84GLIVVSDD673 pKa = 3.85TEE675 pKa = 4.45GSGKK679 pKa = 10.04EE680 pKa = 3.82SAAEE684 pKa = 3.89KK685 pKa = 10.19VSPVRR690 pKa = 11.84PANRR694 pKa = 11.84RR695 pKa = 11.84SIVSVAKK702 pKa = 10.18MIEE705 pKa = 4.0DD706 pKa = 3.0KK707 pKa = 10.38MAEE710 pKa = 3.69VRR712 pKa = 11.84ARR714 pKa = 11.84EE715 pKa = 4.04KK716 pKa = 11.01ARR718 pKa = 11.84TLSPPRR724 pKa = 11.84QTGEE728 pKa = 3.97VEE730 pKa = 3.69ADD732 pKa = 3.06KK733 pKa = 10.26PQQGEE738 pKa = 3.89ADD740 pKa = 3.41TSEE743 pKa = 4.32KK744 pKa = 10.71VDD746 pKa = 3.54VPEE749 pKa = 4.3TKK751 pKa = 10.6EE752 pKa = 3.71MDD754 pKa = 4.09KK755 pKa = 10.78PALPANVAKK764 pKa = 10.16QVSVTMPEE772 pKa = 3.73KK773 pKa = 10.29STEE776 pKa = 3.03KK777 pKa = 9.46WRR779 pKa = 11.84RR780 pKa = 11.84LKK782 pKa = 10.89EE783 pKa = 3.98PTNTQIPDD791 pKa = 3.29EE792 pKa = 4.21FLVDD796 pKa = 3.89NKK798 pKa = 10.86NDD800 pKa = 3.98LKK802 pKa = 10.65WRR804 pKa = 11.84EE805 pKa = 3.76INIDD809 pKa = 3.63KK810 pKa = 10.63EE811 pKa = 4.18NEE813 pKa = 3.72EE814 pKa = 4.66MITMEE819 pKa = 4.7SRR821 pKa = 11.84VEE823 pKa = 3.95QVNEE827 pKa = 3.35ADD829 pKa = 3.51KK830 pKa = 10.8RR831 pKa = 11.84AKK833 pKa = 9.94EE834 pKa = 3.9EE835 pKa = 3.69RR836 pKa = 11.84AEE838 pKa = 4.2RR839 pKa = 11.84VPADD843 pKa = 4.07KK844 pKa = 11.05KK845 pKa = 9.88SDD847 pKa = 3.42AYY849 pKa = 11.24RR850 pKa = 11.84MLVEE854 pKa = 4.8KK855 pKa = 10.91NVDD858 pKa = 3.31PTKK861 pKa = 10.96KK862 pKa = 10.32LFGMSAAKK870 pKa = 9.25WLEE873 pKa = 3.4YY874 pKa = 10.84MMTNKK879 pKa = 10.0PKK881 pKa = 10.49GYY883 pKa = 8.63YY884 pKa = 9.73EE885 pKa = 4.43VMRR888 pKa = 11.84SDD890 pKa = 3.14INEE893 pKa = 4.66FISKK897 pKa = 8.11YY898 pKa = 8.05TATTIAPKK906 pKa = 10.29HH907 pKa = 5.89ILDD910 pKa = 4.0WVPFADD916 pKa = 3.25SVQINGALRR925 pKa = 11.84SARR928 pKa = 11.84RR929 pKa = 11.84SQTSGTNYY937 pKa = 10.15LFSEE941 pKa = 4.34SDD943 pKa = 3.43STIMNVALNIIRR955 pKa = 11.84KK956 pKa = 9.48SGGEE960 pKa = 3.81VWPVSMKK967 pKa = 10.92YY968 pKa = 8.37MGRR971 pKa = 11.84MQRR974 pKa = 11.84EE975 pKa = 4.1FYY977 pKa = 10.27VNKK980 pKa = 9.89VDD982 pKa = 3.45RR983 pKa = 11.84TAYY986 pKa = 10.2NLAAQKK992 pKa = 10.89HH993 pKa = 4.6MM994 pKa = 4.36
Molecular weight: 112.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3642 |
677 |
1087 |
910.5 |
102.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.661 ± 0.372 | 1.593 ± 0.41 |
6.645 ± 0.284 | 7.221 ± 0.307 |
2.773 ± 0.441 | 6.727 ± 0.557 |
1.922 ± 0.427 | 4.283 ± 0.215 |
7.111 ± 0.15 | 7.578 ± 0.338 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.668 ± 0.096 | 3.926 ± 0.243 |
3.597 ± 0.433 | 2.444 ± 0.187 |
6.233 ± 0.589 | 6.233 ± 1.093 |
5.601 ± 0.288 | 7.908 ± 0.233 |
2.004 ± 0.29 | 3.871 ± 0.26 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
