
Hubei diptera virus 9
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sigmavirus; Hubei sigmavirus
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
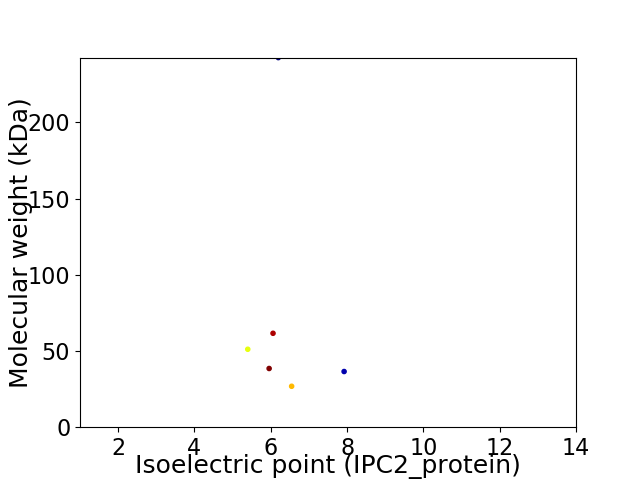
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KMW8|A0A1L3KMW8_9VIRU Putative X protein OS=Hubei diptera virus 9 OX=1922889 PE=4 SV=1
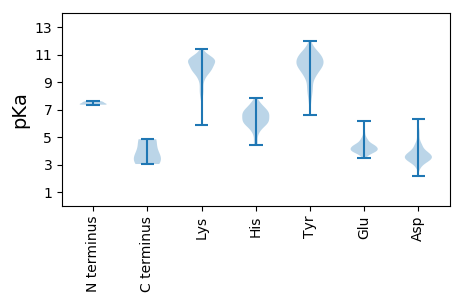
MM1 pKa = 7.55NSLNQTRR8 pKa = 11.84VFCTLDD14 pKa = 3.17GKK16 pKa = 10.61FYY18 pKa = 11.02NVSNVDD24 pKa = 3.44DD25 pKa = 4.43SKK27 pKa = 10.66PTSFPSAWFEE37 pKa = 4.17ANRR40 pKa = 11.84GQKK43 pKa = 9.87PKK45 pKa = 10.86LNIMKK50 pKa = 10.02DD51 pKa = 3.53VPLSKK56 pKa = 10.5LRR58 pKa = 11.84ALMLSSPTNPKK69 pKa = 9.44VPVIAVVTYY78 pKa = 10.05LYY80 pKa = 11.02LVLKK84 pKa = 9.26EE85 pKa = 4.63RR86 pKa = 11.84KK87 pKa = 7.91TICPDD92 pKa = 2.83DD93 pKa = 3.51WSSYY97 pKa = 7.34NTSICSANSQISPFDD112 pKa = 3.5LVTIGEE118 pKa = 4.19IEE120 pKa = 4.77AIEE123 pKa = 4.73LDD125 pKa = 4.0TVVEE129 pKa = 4.13RR130 pKa = 11.84DD131 pKa = 3.75LPSGADD137 pKa = 2.88KK138 pKa = 10.58WIIIMVLSIYY148 pKa = 10.16RR149 pKa = 11.84LEE151 pKa = 4.37RR152 pKa = 11.84ARR154 pKa = 11.84EE155 pKa = 3.76PAYY158 pKa = 10.09EE159 pKa = 3.85ATLIRR164 pKa = 11.84NITNLIKK171 pKa = 10.72ACGCTEE177 pKa = 3.93PPNLTSLLRR186 pKa = 11.84LHH188 pKa = 6.8LKK190 pKa = 8.19WANDD194 pKa = 3.87PGFKK198 pKa = 9.58TAVAAIDD205 pKa = 3.22MFYY208 pKa = 11.14YY209 pKa = 10.51RR210 pKa = 11.84FEE212 pKa = 4.3NDD214 pKa = 2.18EE215 pKa = 3.9WADD218 pKa = 3.25IRR220 pKa = 11.84IATIGSRR227 pKa = 11.84LKK229 pKa = 10.72DD230 pKa = 3.42CSAIQSLTQTASIAGLSSTHH250 pKa = 6.74EE251 pKa = 3.98IIYY254 pKa = 9.65WIGNVAVGTEE264 pKa = 4.13FSKK267 pKa = 10.53MSSSVEE273 pKa = 4.01EE274 pKa = 4.08AGMLYY279 pKa = 10.23SYY281 pKa = 10.71FPYY284 pKa = 10.4QVDD287 pKa = 4.27FGLVTRR293 pKa = 11.84SAYY296 pKa = 9.06SARR299 pKa = 11.84VNPALHH305 pKa = 6.05TWLHH309 pKa = 6.06IIGIFFCNSRR319 pKa = 11.84SLNAKK324 pKa = 9.66FFLEE328 pKa = 3.95KK329 pKa = 10.59DD330 pKa = 3.47VAGVIRR336 pKa = 11.84SAQWVGYY343 pKa = 8.33VLSRR347 pKa = 11.84GQHH350 pKa = 4.91SGRR353 pKa = 11.84IFVPADD359 pKa = 3.58RR360 pKa = 11.84PEE362 pKa = 3.81QDD364 pKa = 5.43MVDD367 pKa = 3.47EE368 pKa = 4.44WNEE371 pKa = 3.95NQQEE375 pKa = 4.12ATDD378 pKa = 3.97ANEE381 pKa = 4.09GVEE384 pKa = 4.66DD385 pKa = 4.02LTLGAISDD393 pKa = 3.93TDD395 pKa = 3.65LTRR398 pKa = 11.84EE399 pKa = 4.07PLSNNPFEE407 pKa = 4.38WFSYY411 pKa = 9.29MSHH414 pKa = 7.1RR415 pKa = 11.84DD416 pKa = 3.6YY417 pKa = 11.6KK418 pKa = 9.88LTQSMRR424 pKa = 11.84SKK426 pKa = 10.1IRR428 pKa = 11.84SVQSEE433 pKa = 4.28IVNPRR438 pKa = 11.84PGTIAAMAQTSLFYY452 pKa = 11.07HH453 pKa = 6.78EE454 pKa = 4.83
MM1 pKa = 7.55NSLNQTRR8 pKa = 11.84VFCTLDD14 pKa = 3.17GKK16 pKa = 10.61FYY18 pKa = 11.02NVSNVDD24 pKa = 3.44DD25 pKa = 4.43SKK27 pKa = 10.66PTSFPSAWFEE37 pKa = 4.17ANRR40 pKa = 11.84GQKK43 pKa = 9.87PKK45 pKa = 10.86LNIMKK50 pKa = 10.02DD51 pKa = 3.53VPLSKK56 pKa = 10.5LRR58 pKa = 11.84ALMLSSPTNPKK69 pKa = 9.44VPVIAVVTYY78 pKa = 10.05LYY80 pKa = 11.02LVLKK84 pKa = 9.26EE85 pKa = 4.63RR86 pKa = 11.84KK87 pKa = 7.91TICPDD92 pKa = 2.83DD93 pKa = 3.51WSSYY97 pKa = 7.34NTSICSANSQISPFDD112 pKa = 3.5LVTIGEE118 pKa = 4.19IEE120 pKa = 4.77AIEE123 pKa = 4.73LDD125 pKa = 4.0TVVEE129 pKa = 4.13RR130 pKa = 11.84DD131 pKa = 3.75LPSGADD137 pKa = 2.88KK138 pKa = 10.58WIIIMVLSIYY148 pKa = 10.16RR149 pKa = 11.84LEE151 pKa = 4.37RR152 pKa = 11.84ARR154 pKa = 11.84EE155 pKa = 3.76PAYY158 pKa = 10.09EE159 pKa = 3.85ATLIRR164 pKa = 11.84NITNLIKK171 pKa = 10.72ACGCTEE177 pKa = 3.93PPNLTSLLRR186 pKa = 11.84LHH188 pKa = 6.8LKK190 pKa = 8.19WANDD194 pKa = 3.87PGFKK198 pKa = 9.58TAVAAIDD205 pKa = 3.22MFYY208 pKa = 11.14YY209 pKa = 10.51RR210 pKa = 11.84FEE212 pKa = 4.3NDD214 pKa = 2.18EE215 pKa = 3.9WADD218 pKa = 3.25IRR220 pKa = 11.84IATIGSRR227 pKa = 11.84LKK229 pKa = 10.72DD230 pKa = 3.42CSAIQSLTQTASIAGLSSTHH250 pKa = 6.74EE251 pKa = 3.98IIYY254 pKa = 9.65WIGNVAVGTEE264 pKa = 4.13FSKK267 pKa = 10.53MSSSVEE273 pKa = 4.01EE274 pKa = 4.08AGMLYY279 pKa = 10.23SYY281 pKa = 10.71FPYY284 pKa = 10.4QVDD287 pKa = 4.27FGLVTRR293 pKa = 11.84SAYY296 pKa = 9.06SARR299 pKa = 11.84VNPALHH305 pKa = 6.05TWLHH309 pKa = 6.06IIGIFFCNSRR319 pKa = 11.84SLNAKK324 pKa = 9.66FFLEE328 pKa = 3.95KK329 pKa = 10.59DD330 pKa = 3.47VAGVIRR336 pKa = 11.84SAQWVGYY343 pKa = 8.33VLSRR347 pKa = 11.84GQHH350 pKa = 4.91SGRR353 pKa = 11.84IFVPADD359 pKa = 3.58RR360 pKa = 11.84PEE362 pKa = 3.81QDD364 pKa = 5.43MVDD367 pKa = 3.47EE368 pKa = 4.44WNEE371 pKa = 3.95NQQEE375 pKa = 4.12ATDD378 pKa = 3.97ANEE381 pKa = 4.09GVEE384 pKa = 4.66DD385 pKa = 4.02LTLGAISDD393 pKa = 3.93TDD395 pKa = 3.65LTRR398 pKa = 11.84EE399 pKa = 4.07PLSNNPFEE407 pKa = 4.38WFSYY411 pKa = 9.29MSHH414 pKa = 7.1RR415 pKa = 11.84DD416 pKa = 3.6YY417 pKa = 11.6KK418 pKa = 9.88LTQSMRR424 pKa = 11.84SKK426 pKa = 10.1IRR428 pKa = 11.84SVQSEE433 pKa = 4.28IVNPRR438 pKa = 11.84PGTIAAMAQTSLFYY452 pKa = 11.07HH453 pKa = 6.78EE454 pKa = 4.83
Molecular weight: 51.15 kDa
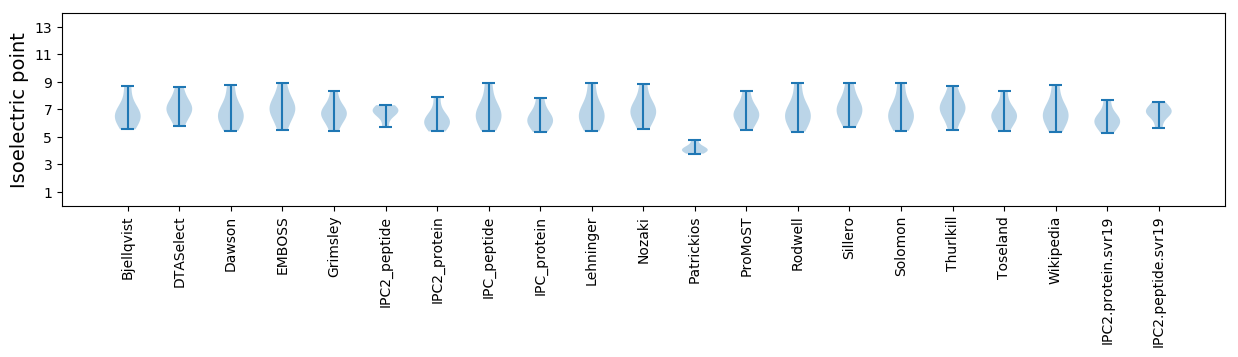
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KMU2|A0A1L3KMU2_9VIRU Nucleocapsid protein OS=Hubei diptera virus 9 OX=1922889 PE=4 SV=1
MM1 pKa = 7.49KK2 pKa = 10.2KK3 pKa = 10.31INMEE7 pKa = 4.31NNNTTKK13 pKa = 10.08MNKK16 pKa = 8.14KK17 pKa = 8.16TPNKK21 pKa = 9.1SQSKK25 pKa = 7.99TRR27 pKa = 11.84STKK30 pKa = 9.09QRR32 pKa = 11.84EE33 pKa = 4.23INPPLLQQFGPNTPQDD49 pKa = 3.52PKK51 pKa = 11.11KK52 pKa = 10.03WFDD55 pKa = 3.49EE56 pKa = 4.43AQSRR60 pKa = 11.84PGEE63 pKa = 3.94MLEE66 pKa = 4.5NVMKK70 pKa = 10.03TLAKK74 pKa = 10.53DD75 pKa = 3.88GPNPEE80 pKa = 3.94AEE82 pKa = 4.06IRR84 pKa = 11.84EE85 pKa = 4.12EE86 pKa = 4.9DD87 pKa = 4.52SGFLNPNIPTGIIDD101 pKa = 5.17DD102 pKa = 5.49PIEE105 pKa = 4.39CMAVGAAALHH115 pKa = 6.04LSEE118 pKa = 5.98FPTTSKK124 pKa = 10.58RR125 pKa = 11.84GRR127 pKa = 11.84EE128 pKa = 3.84VNTYY132 pKa = 8.15KK133 pKa = 10.33TAEE136 pKa = 3.79THH138 pKa = 6.25QYY140 pKa = 7.91VTADD144 pKa = 4.07LIEE147 pKa = 4.63NKK149 pKa = 9.48FQNGQSYY156 pKa = 11.27DD157 pKa = 3.27SDD159 pKa = 3.81NFLMIFKK166 pKa = 10.42KK167 pKa = 10.74LKK169 pKa = 9.82TNMGDD174 pKa = 3.27TADD177 pKa = 4.41KK178 pKa = 10.85YY179 pKa = 11.1FKK181 pKa = 9.94LTEE184 pKa = 4.56GEE186 pKa = 4.45GEE188 pKa = 3.95WLITLKK194 pKa = 10.77NPYY197 pKa = 9.32PSPEE201 pKa = 4.45LKK203 pKa = 10.2RR204 pKa = 11.84QRR206 pKa = 11.84QTEE209 pKa = 4.39CIPTSSKK216 pKa = 5.89QTKK219 pKa = 10.25VIFPDD224 pKa = 3.29EE225 pKa = 4.16NTNQGASEE233 pKa = 4.33TEE235 pKa = 3.88IVNPLLEE242 pKa = 4.96SYY244 pKa = 11.02LKK246 pKa = 10.72FLTPGLTLKK255 pKa = 10.62SRR257 pKa = 11.84EE258 pKa = 4.06YY259 pKa = 11.41GLGSLKK265 pKa = 10.35ISLGSFSMRR274 pKa = 11.84EE275 pKa = 3.67EE276 pKa = 3.8DD277 pKa = 3.29VRR279 pKa = 11.84AYY281 pKa = 10.3FVGKK285 pKa = 9.86EE286 pKa = 3.57IPTTFRR292 pKa = 11.84AFVYY296 pKa = 9.75TLAKK300 pKa = 9.35CQPKK304 pKa = 10.0IIRR307 pKa = 11.84VLNKK311 pKa = 10.17YY312 pKa = 9.46IVPLNVKK319 pKa = 8.77STDD322 pKa = 3.04
MM1 pKa = 7.49KK2 pKa = 10.2KK3 pKa = 10.31INMEE7 pKa = 4.31NNNTTKK13 pKa = 10.08MNKK16 pKa = 8.14KK17 pKa = 8.16TPNKK21 pKa = 9.1SQSKK25 pKa = 7.99TRR27 pKa = 11.84STKK30 pKa = 9.09QRR32 pKa = 11.84EE33 pKa = 4.23INPPLLQQFGPNTPQDD49 pKa = 3.52PKK51 pKa = 11.11KK52 pKa = 10.03WFDD55 pKa = 3.49EE56 pKa = 4.43AQSRR60 pKa = 11.84PGEE63 pKa = 3.94MLEE66 pKa = 4.5NVMKK70 pKa = 10.03TLAKK74 pKa = 10.53DD75 pKa = 3.88GPNPEE80 pKa = 3.94AEE82 pKa = 4.06IRR84 pKa = 11.84EE85 pKa = 4.12EE86 pKa = 4.9DD87 pKa = 4.52SGFLNPNIPTGIIDD101 pKa = 5.17DD102 pKa = 5.49PIEE105 pKa = 4.39CMAVGAAALHH115 pKa = 6.04LSEE118 pKa = 5.98FPTTSKK124 pKa = 10.58RR125 pKa = 11.84GRR127 pKa = 11.84EE128 pKa = 3.84VNTYY132 pKa = 8.15KK133 pKa = 10.33TAEE136 pKa = 3.79THH138 pKa = 6.25QYY140 pKa = 7.91VTADD144 pKa = 4.07LIEE147 pKa = 4.63NKK149 pKa = 9.48FQNGQSYY156 pKa = 11.27DD157 pKa = 3.27SDD159 pKa = 3.81NFLMIFKK166 pKa = 10.42KK167 pKa = 10.74LKK169 pKa = 9.82TNMGDD174 pKa = 3.27TADD177 pKa = 4.41KK178 pKa = 10.85YY179 pKa = 11.1FKK181 pKa = 9.94LTEE184 pKa = 4.56GEE186 pKa = 4.45GEE188 pKa = 3.95WLITLKK194 pKa = 10.77NPYY197 pKa = 9.32PSPEE201 pKa = 4.45LKK203 pKa = 10.2RR204 pKa = 11.84QRR206 pKa = 11.84QTEE209 pKa = 4.39CIPTSSKK216 pKa = 5.89QTKK219 pKa = 10.25VIFPDD224 pKa = 3.29EE225 pKa = 4.16NTNQGASEE233 pKa = 4.33TEE235 pKa = 3.88IVNPLLEE242 pKa = 4.96SYY244 pKa = 11.02LKK246 pKa = 10.72FLTPGLTLKK255 pKa = 10.62SRR257 pKa = 11.84EE258 pKa = 4.06YY259 pKa = 11.41GLGSLKK265 pKa = 10.35ISLGSFSMRR274 pKa = 11.84EE275 pKa = 3.67EE276 pKa = 3.8DD277 pKa = 3.29VRR279 pKa = 11.84AYY281 pKa = 10.3FVGKK285 pKa = 9.86EE286 pKa = 3.57IPTTFRR292 pKa = 11.84AFVYY296 pKa = 9.75TLAKK300 pKa = 9.35CQPKK304 pKa = 10.0IIRR307 pKa = 11.84VLNKK311 pKa = 10.17YY312 pKa = 9.46IVPLNVKK319 pKa = 8.77STDD322 pKa = 3.04
Molecular weight: 36.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3998 |
229 |
2120 |
666.3 |
76.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.977 ± 0.701 | 1.501 ± 0.184 |
5.078 ± 0.168 | 6.203 ± 0.244 |
4.677 ± 0.215 | 4.852 ± 0.201 |
2.551 ± 0.47 | 7.529 ± 0.63 |
5.628 ± 0.501 | 10.63 ± 1.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.701 ± 0.185 | 6.053 ± 0.208 |
5.078 ± 0.307 | 3.352 ± 0.235 |
5.078 ± 0.276 | 8.579 ± 0.388 |
6.428 ± 0.468 | 5.603 ± 0.357 |
1.726 ± 0.143 | 3.777 ± 0.399 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
