
Cutthroat trout virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Hepelivirales; Hepeviridae; Piscihepevirus; Piscihepevirus A
Average proteome isoelectric point is 7.22
Get precalculated fractions of proteins
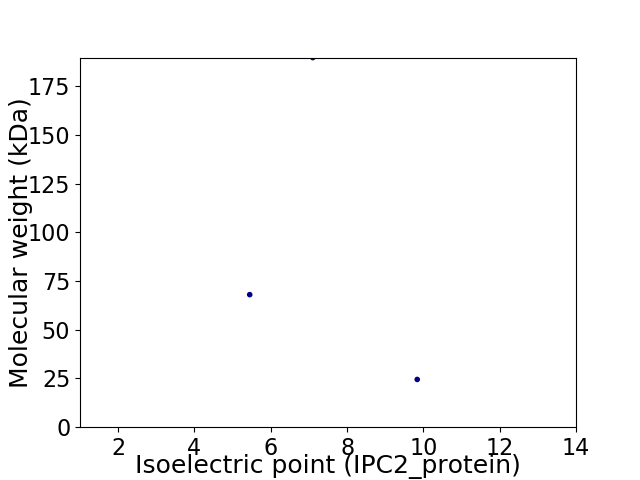
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F5BB19|F5BB19_9VIRU Phosphoprotein OS=Cutthroat trout virus OX=1016879 PE=4 SV=1
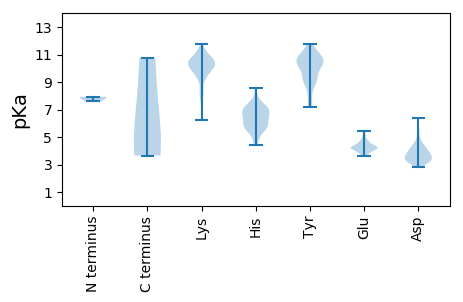
MM1 pKa = 7.85ADD3 pKa = 3.24SNAVVPSAQSKK14 pKa = 8.73PRR16 pKa = 11.84RR17 pKa = 11.84TRR19 pKa = 11.84RR20 pKa = 11.84PKK22 pKa = 9.58MDD24 pKa = 4.35PSQIPDD30 pKa = 3.47TVKK33 pKa = 9.99TFRR36 pKa = 11.84LTTDD40 pKa = 5.14PIMAPPPGEE49 pKa = 4.13IDD51 pKa = 3.42SLLYY55 pKa = 10.47TSGLSPAVVLSFSSFTSPAVVFAANYY81 pKa = 7.29EE82 pKa = 4.28KK83 pKa = 11.3YY84 pKa = 10.51KK85 pKa = 10.54ILSFKK90 pKa = 10.43ATVLSLAPEE99 pKa = 5.18AISSYY104 pKa = 10.13NLSVVWFPDD113 pKa = 3.15ASEE116 pKa = 5.15GPTSLQATNLSMVRR130 pKa = 11.84RR131 pKa = 11.84YY132 pKa = 10.28HH133 pKa = 7.13SILPGQSGSLSLTASEE149 pKa = 4.5LLHH152 pKa = 6.91GIAEE156 pKa = 4.49KK157 pKa = 11.0YY158 pKa = 10.29VDD160 pKa = 4.1PTHH163 pKa = 7.1SDD165 pKa = 3.11ALSAFCGLFMVVCHH179 pKa = 6.19GAPFNHH185 pKa = 5.94YY186 pKa = 10.33RR187 pKa = 11.84DD188 pKa = 3.88TQYY191 pKa = 11.35LGPLFKK197 pKa = 10.53IQFEE201 pKa = 4.48FKK203 pKa = 10.63LSVFGYY209 pKa = 9.8SPGNIPVKK217 pKa = 10.54VQGASATEE225 pKa = 3.9IVSFGTVDD233 pKa = 3.58GKK235 pKa = 10.23ATFDD239 pKa = 3.68TPSAIALHH247 pKa = 6.79ASYY250 pKa = 10.96QSGPPNSPGPSIGQVLLTVADD271 pKa = 4.16TVSAALPFPFSILAKK286 pKa = 10.13VGLFFIRR293 pKa = 11.84KK294 pKa = 6.7ITGANGTSFMVYY306 pKa = 9.99KK307 pKa = 10.57SYY309 pKa = 11.74GDD311 pKa = 4.23AIRR314 pKa = 11.84NRR316 pKa = 11.84PLPSRR321 pKa = 11.84SSHH324 pKa = 4.73QLGNALVRR332 pKa = 11.84QTTFGTNDD340 pKa = 3.68DD341 pKa = 4.36SVLPPQPISQFTFHH355 pKa = 7.23FKK357 pKa = 10.32TGDD360 pKa = 3.35VLVIQGSFKK369 pKa = 11.16APTSRR374 pKa = 11.84PNNWNVQEE382 pKa = 4.25FSISRR387 pKa = 11.84ILNITNGTSTTNILVFEE404 pKa = 4.22KK405 pKa = 8.79WTLDD409 pKa = 3.31GNLPQIKK416 pKa = 8.3NTYY419 pKa = 9.17GLIGVHH425 pKa = 5.8YY426 pKa = 9.29FPASISPVGTSTFDD440 pKa = 4.77NIIGDD445 pKa = 4.14CKK447 pKa = 10.91VFAGTATDD455 pKa = 3.8SMLTPGPGSITVTFLSQTRR474 pKa = 11.84GFLSQTEE481 pKa = 4.25LTQGLIAASSTMDD494 pKa = 3.35TTDD497 pKa = 3.71SSLTRR502 pKa = 11.84QASQPMEE509 pKa = 3.75VDD511 pKa = 3.55GFLSGLIPSTPLCNMVNNTFSCGNSDD537 pKa = 3.35SQATSRR543 pKa = 11.84SAVHH547 pKa = 6.51SLGPNCDD554 pKa = 3.03KK555 pKa = 11.28CGLDD559 pKa = 3.29LNLRR563 pKa = 11.84VTPCIATRR571 pKa = 11.84LQHH574 pKa = 5.66TPKK577 pKa = 10.49ACPHH581 pKa = 6.35CSFLLGLTAYY591 pKa = 8.96TVDD594 pKa = 3.95CMRR597 pKa = 11.84PSDD600 pKa = 4.0TTDD603 pKa = 3.06STVDD607 pKa = 3.23ALLNPLNSLDD617 pKa = 4.5LEE619 pKa = 4.68SEE621 pKa = 4.44SEE623 pKa = 4.13PSDD626 pKa = 4.18DD627 pKa = 6.36DD628 pKa = 5.97LIQFDD633 pKa = 4.35DD634 pKa = 3.66
MM1 pKa = 7.85ADD3 pKa = 3.24SNAVVPSAQSKK14 pKa = 8.73PRR16 pKa = 11.84RR17 pKa = 11.84TRR19 pKa = 11.84RR20 pKa = 11.84PKK22 pKa = 9.58MDD24 pKa = 4.35PSQIPDD30 pKa = 3.47TVKK33 pKa = 9.99TFRR36 pKa = 11.84LTTDD40 pKa = 5.14PIMAPPPGEE49 pKa = 4.13IDD51 pKa = 3.42SLLYY55 pKa = 10.47TSGLSPAVVLSFSSFTSPAVVFAANYY81 pKa = 7.29EE82 pKa = 4.28KK83 pKa = 11.3YY84 pKa = 10.51KK85 pKa = 10.54ILSFKK90 pKa = 10.43ATVLSLAPEE99 pKa = 5.18AISSYY104 pKa = 10.13NLSVVWFPDD113 pKa = 3.15ASEE116 pKa = 5.15GPTSLQATNLSMVRR130 pKa = 11.84RR131 pKa = 11.84YY132 pKa = 10.28HH133 pKa = 7.13SILPGQSGSLSLTASEE149 pKa = 4.5LLHH152 pKa = 6.91GIAEE156 pKa = 4.49KK157 pKa = 11.0YY158 pKa = 10.29VDD160 pKa = 4.1PTHH163 pKa = 7.1SDD165 pKa = 3.11ALSAFCGLFMVVCHH179 pKa = 6.19GAPFNHH185 pKa = 5.94YY186 pKa = 10.33RR187 pKa = 11.84DD188 pKa = 3.88TQYY191 pKa = 11.35LGPLFKK197 pKa = 10.53IQFEE201 pKa = 4.48FKK203 pKa = 10.63LSVFGYY209 pKa = 9.8SPGNIPVKK217 pKa = 10.54VQGASATEE225 pKa = 3.9IVSFGTVDD233 pKa = 3.58GKK235 pKa = 10.23ATFDD239 pKa = 3.68TPSAIALHH247 pKa = 6.79ASYY250 pKa = 10.96QSGPPNSPGPSIGQVLLTVADD271 pKa = 4.16TVSAALPFPFSILAKK286 pKa = 10.13VGLFFIRR293 pKa = 11.84KK294 pKa = 6.7ITGANGTSFMVYY306 pKa = 9.99KK307 pKa = 10.57SYY309 pKa = 11.74GDD311 pKa = 4.23AIRR314 pKa = 11.84NRR316 pKa = 11.84PLPSRR321 pKa = 11.84SSHH324 pKa = 4.73QLGNALVRR332 pKa = 11.84QTTFGTNDD340 pKa = 3.68DD341 pKa = 4.36SVLPPQPISQFTFHH355 pKa = 7.23FKK357 pKa = 10.32TGDD360 pKa = 3.35VLVIQGSFKK369 pKa = 11.16APTSRR374 pKa = 11.84PNNWNVQEE382 pKa = 4.25FSISRR387 pKa = 11.84ILNITNGTSTTNILVFEE404 pKa = 4.22KK405 pKa = 8.79WTLDD409 pKa = 3.31GNLPQIKK416 pKa = 8.3NTYY419 pKa = 9.17GLIGVHH425 pKa = 5.8YY426 pKa = 9.29FPASISPVGTSTFDD440 pKa = 4.77NIIGDD445 pKa = 4.14CKK447 pKa = 10.91VFAGTATDD455 pKa = 3.8SMLTPGPGSITVTFLSQTRR474 pKa = 11.84GFLSQTEE481 pKa = 4.25LTQGLIAASSTMDD494 pKa = 3.35TTDD497 pKa = 3.71SSLTRR502 pKa = 11.84QASQPMEE509 pKa = 3.75VDD511 pKa = 3.55GFLSGLIPSTPLCNMVNNTFSCGNSDD537 pKa = 3.35SQATSRR543 pKa = 11.84SAVHH547 pKa = 6.51SLGPNCDD554 pKa = 3.03KK555 pKa = 11.28CGLDD559 pKa = 3.29LNLRR563 pKa = 11.84VTPCIATRR571 pKa = 11.84LQHH574 pKa = 5.66TPKK577 pKa = 10.49ACPHH581 pKa = 6.35CSFLLGLTAYY591 pKa = 8.96TVDD594 pKa = 3.95CMRR597 pKa = 11.84PSDD600 pKa = 4.0TTDD603 pKa = 3.06STVDD607 pKa = 3.23ALLNPLNSLDD617 pKa = 4.5LEE619 pKa = 4.68SEE621 pKa = 4.44SEE623 pKa = 4.13PSDD626 pKa = 4.18DD627 pKa = 6.36DD628 pKa = 5.97LIQFDD633 pKa = 4.35DD634 pKa = 3.66
Molecular weight: 68.01 kDa
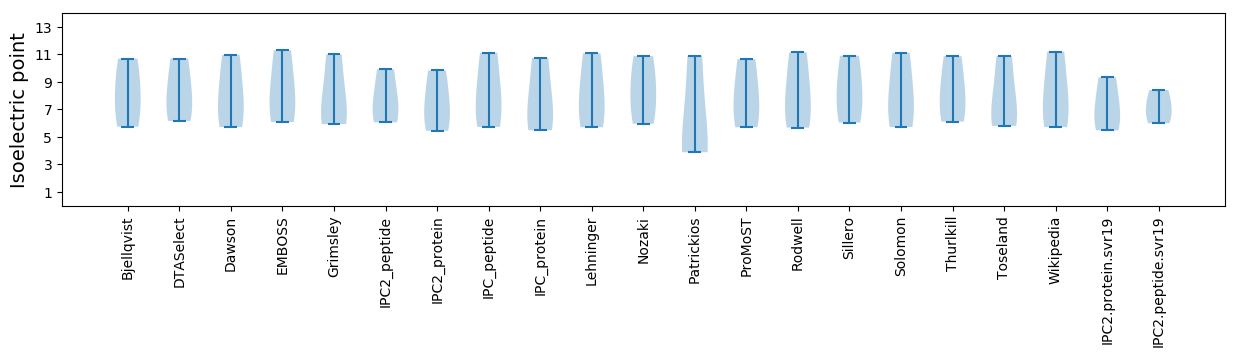
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F5BB19|F5BB19_9VIRU Phosphoprotein OS=Cutthroat trout virus OX=1016879 PE=4 SV=1
MM1 pKa = 7.59GKK3 pKa = 10.07LPLTPLPQLLSMLHH17 pKa = 5.86TKK19 pKa = 9.56VAHH22 pKa = 5.97QIHH25 pKa = 7.15PGLLSAKK32 pKa = 10.13YY33 pKa = 9.58SSQWLTPCQLRR44 pKa = 11.84SLSLSVYY51 pKa = 8.98LQKK54 pKa = 10.92LVSSLSARR62 pKa = 11.84LLEE65 pKa = 4.28PMEE68 pKa = 4.38LPSWSTNPMEE78 pKa = 4.34MQFATAHH85 pKa = 5.8YY86 pKa = 9.08PHH88 pKa = 7.0AQAINWAMHH97 pKa = 6.21LSARR101 pKa = 11.84PHH103 pKa = 6.17SALTMTQFSHH113 pKa = 7.35PSPSASSPSTSRR125 pKa = 11.84LAMSLSSRR133 pKa = 11.84VPSRR137 pKa = 11.84PQPVDD142 pKa = 3.64PIIGMFRR149 pKa = 11.84NSPFRR154 pKa = 11.84EE155 pKa = 4.19SLTSLMGPPLQTFLCLKK172 pKa = 9.86SGLWMVTSHH181 pKa = 7.51RR182 pKa = 11.84SRR184 pKa = 11.84TPTVSSGSTTSRR196 pKa = 11.84LPLVLLAPAPSTTSLVIAKK215 pKa = 9.8SLQAPQLTACC225 pKa = 4.68
MM1 pKa = 7.59GKK3 pKa = 10.07LPLTPLPQLLSMLHH17 pKa = 5.86TKK19 pKa = 9.56VAHH22 pKa = 5.97QIHH25 pKa = 7.15PGLLSAKK32 pKa = 10.13YY33 pKa = 9.58SSQWLTPCQLRR44 pKa = 11.84SLSLSVYY51 pKa = 8.98LQKK54 pKa = 10.92LVSSLSARR62 pKa = 11.84LLEE65 pKa = 4.28PMEE68 pKa = 4.38LPSWSTNPMEE78 pKa = 4.34MQFATAHH85 pKa = 5.8YY86 pKa = 9.08PHH88 pKa = 7.0AQAINWAMHH97 pKa = 6.21LSARR101 pKa = 11.84PHH103 pKa = 6.17SALTMTQFSHH113 pKa = 7.35PSPSASSPSTSRR125 pKa = 11.84LAMSLSSRR133 pKa = 11.84VPSRR137 pKa = 11.84PQPVDD142 pKa = 3.64PIIGMFRR149 pKa = 11.84NSPFRR154 pKa = 11.84EE155 pKa = 4.19SLTSLMGPPLQTFLCLKK172 pKa = 9.86SGLWMVTSHH181 pKa = 7.51RR182 pKa = 11.84SRR184 pKa = 11.84TPTVSSGSTTSRR196 pKa = 11.84LPLVLLAPAPSTTSLVIAKK215 pKa = 9.8SLQAPQLTACC225 pKa = 4.68
Molecular weight: 24.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2566 |
225 |
1707 |
855.3 |
94.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.093 ± 0.116 | 1.949 ± 0.212 |
5.222 ± 1.224 | 3.468 ± 0.742 |
4.287 ± 0.626 | 5.417 ± 0.718 |
3.118 ± 0.462 | 5.222 ± 0.832 |
4.482 ± 0.792 | 9.314 ± 1.695 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.027 ± 0.732 | 4.092 ± 0.701 |
7.443 ± 1.03 | 4.092 ± 0.309 |
4.521 ± 0.46 | 9.275 ± 2.644 |
9.353 ± 0.4 | 5.924 ± 0.48 |
0.935 ± 0.234 | 2.767 ± 0.468 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
