
Solemya pervernicosa gill symbiont
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Gammaproteobacteria incertae sedis; sulfur-oxidizing symbionts
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
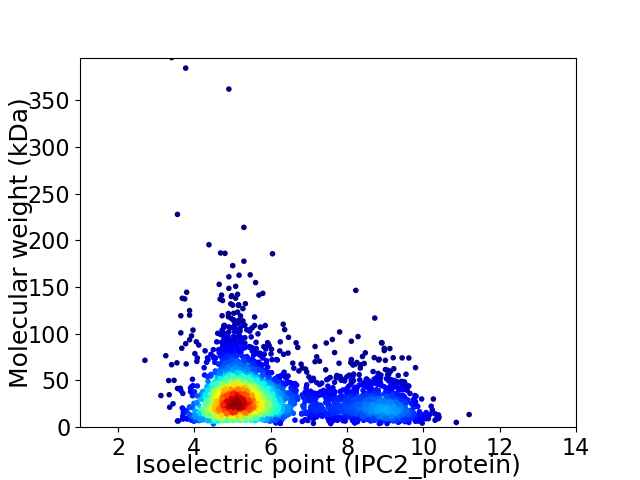
Virtual 2D-PAGE plot for 3236 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
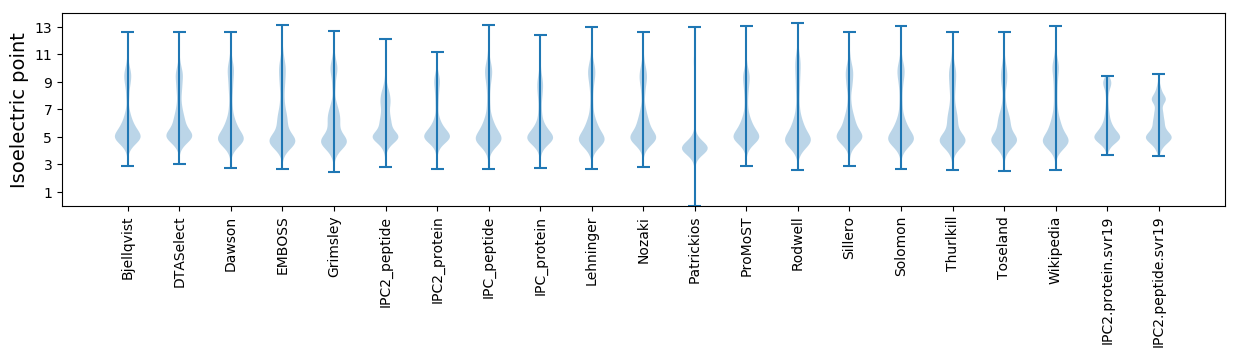
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1T2L373|A0A1T2L373_9GAMM Probable chromosome-partitioning protein ParB OS=Solemya pervernicosa gill symbiont OX=642797 GN=BOW53_11815 PE=3 SV=1
MM1 pKa = 6.96STNEE5 pKa = 4.0APSASFIVDD14 pKa = 3.39VGTGHH19 pKa = 6.61SPLMVNFDD27 pKa = 4.56ASPSSDD33 pKa = 3.07SDD35 pKa = 3.71GSIAGYY41 pKa = 9.92SWLFGDD47 pKa = 5.12GSTATGQQVSHH58 pKa = 6.98SYY60 pKa = 10.47TNVGNHH66 pKa = 5.5SVTLTVTDD74 pKa = 3.57NEE76 pKa = 4.4GAIDD80 pKa = 3.86NATPQTISVWIANNAPTALFSVDD103 pKa = 3.28TDD105 pKa = 3.64SGEE108 pKa = 4.36GPLTVNFDD116 pKa = 3.52ASASGDD122 pKa = 3.45SDD124 pKa = 3.75GTIVAYY130 pKa = 9.89GWDD133 pKa = 3.87FGDD136 pKa = 3.96GSSGNGQTVAHH147 pKa = 7.09SYY149 pKa = 6.39TTPGVYY155 pKa = 10.2LATLTVRR162 pKa = 11.84DD163 pKa = 4.28DD164 pKa = 3.91DD165 pKa = 4.52AATKK169 pKa = 10.65LSTAHH174 pKa = 7.71IIRR177 pKa = 11.84VLPANQAPTAAFSVDD192 pKa = 3.45TDD194 pKa = 3.51SGTTPLTVNFDD205 pKa = 3.69ASGSTDD211 pKa = 3.11TDD213 pKa = 3.8GSIVSYY219 pKa = 10.76AWDD222 pKa = 4.21FGDD225 pKa = 4.03GDD227 pKa = 4.16SGVGQQVSHH236 pKa = 7.16TYY238 pKa = 10.08TSDD241 pKa = 2.97GSYY244 pKa = 9.36TATLTVTDD252 pKa = 3.73NAGAEE257 pKa = 4.27DD258 pKa = 4.24SDD260 pKa = 4.47SDD262 pKa = 4.45VIDD265 pKa = 4.76SISASHH271 pKa = 6.49YY272 pKa = 6.45PTARR276 pKa = 11.84YY277 pKa = 9.51DD278 pKa = 3.41VDD280 pKa = 3.9VDD282 pKa = 3.97SGPAPLTVSFDD293 pKa = 3.6ASTSSDD299 pKa = 3.65PDD301 pKa = 3.24NDD303 pKa = 3.71IINYY307 pKa = 8.04FWQFGDD313 pKa = 3.55GDD315 pKa = 3.98NASGVTTSHH324 pKa = 7.15TYY326 pKa = 8.4TVPGSYY332 pKa = 10.0DD333 pKa = 3.49VQMLATDD340 pKa = 3.86SASQGDD346 pKa = 4.08VADD349 pKa = 3.55VRR351 pKa = 11.84TITVQGPNTLPTAAFTVDD369 pKa = 3.77VNSGTSPVTVNFDD382 pKa = 3.2ATTSSDD388 pKa = 2.96SDD390 pKa = 3.96GSVVGYY396 pKa = 10.19GWDD399 pKa = 3.7FGDD402 pKa = 4.42GFQGSGASTTHH413 pKa = 6.62TYY415 pKa = 10.2FADD418 pKa = 4.17GTHH421 pKa = 5.75TAILTVTDD429 pKa = 3.95DD430 pKa = 3.53RR431 pKa = 11.84GGQHH435 pKa = 6.09SVSHH439 pKa = 7.35DD440 pKa = 3.09ISVAASGGVTLSGVLGLSGTGTVDD464 pKa = 4.25SDD466 pKa = 4.22VNDD469 pKa = 3.41INADD473 pKa = 3.66YY474 pKa = 10.94QSNNTTATAQLLISPSSLGNVWGFVSKK501 pKa = 9.19TATGIEE507 pKa = 4.44GEE509 pKa = 4.27RR510 pKa = 11.84FTYY513 pKa = 10.52SADD516 pKa = 3.32EE517 pKa = 3.94VDD519 pKa = 3.91IYY521 pKa = 10.88RR522 pKa = 11.84AYY524 pKa = 10.93LPVGQRR530 pKa = 11.84ICLTIKK536 pKa = 10.87NPDD539 pKa = 3.79DD540 pKa = 3.54EE541 pKa = 5.02TGLEE545 pKa = 4.08NANDD549 pKa = 4.13LDD551 pKa = 4.49LYY553 pKa = 10.61LYY555 pKa = 10.91SVDD558 pKa = 4.88DD559 pKa = 4.23PEE561 pKa = 5.91SPVATALGTHH571 pKa = 5.89SSEE574 pKa = 4.3TLHH577 pKa = 6.4VPTGGEE583 pKa = 4.07YY584 pKa = 10.19YY585 pKa = 10.08IVVAAYY591 pKa = 9.97RR592 pKa = 11.84GKK594 pKa = 8.98STYY597 pKa = 10.95KK598 pKa = 10.46LATFMDD604 pKa = 4.55LTSVDD609 pKa = 3.6EE610 pKa = 4.45VQTLRR615 pKa = 11.84LEE617 pKa = 4.53DD618 pKa = 4.31DD619 pKa = 4.44FVPGQVIAKK628 pKa = 9.53LKK630 pKa = 10.54SEE632 pKa = 4.3VVNPPGAGADD642 pKa = 3.58KK643 pKa = 10.71KK644 pKa = 11.11GVAAATAIEE653 pKa = 4.57SVSSRR658 pKa = 11.84GSVSMIRR665 pKa = 11.84GAADD669 pKa = 3.64RR670 pKa = 11.84ASLFSVNDD678 pKa = 3.3MGQNSASGKK687 pKa = 8.53LQSAASSDD695 pKa = 3.65QDD697 pKa = 3.25EE698 pKa = 4.85SEE700 pKa = 4.15YY701 pKa = 11.53SLLDD705 pKa = 3.11GRR707 pKa = 11.84YY708 pKa = 10.32GIGLSQKK715 pKa = 10.26DD716 pKa = 3.26RR717 pKa = 11.84EE718 pKa = 4.34AFEE721 pKa = 4.3TIKK724 pKa = 10.69AIKK727 pKa = 10.03RR728 pKa = 11.84LRR730 pKa = 11.84QDD732 pKa = 2.64SRR734 pKa = 11.84IDD736 pKa = 3.56YY737 pKa = 10.83AEE739 pKa = 4.12PNYY742 pKa = 8.52IQKK745 pKa = 9.74MSLVPDD751 pKa = 4.34DD752 pKa = 4.45LHH754 pKa = 8.86YY755 pKa = 10.17EE756 pKa = 4.35QQWHH760 pKa = 5.72YY761 pKa = 10.35PAINLPDD768 pKa = 3.17TWDD771 pKa = 3.26ITTGDD776 pKa = 3.37ASVIVAVIDD785 pKa = 3.52SGVFMAHH792 pKa = 6.89EE793 pKa = 4.67DD794 pKa = 3.45LSGNLLSTGYY804 pKa = 10.77DD805 pKa = 3.88FIVSTSMSNDD815 pKa = 2.87GDD817 pKa = 4.88GIDD820 pKa = 5.14PDD822 pKa = 5.2PDD824 pKa = 4.6PDD826 pKa = 4.13DD827 pKa = 5.11PGDD830 pKa = 3.6IVISYY835 pKa = 7.69SVWHH839 pKa = 5.97GTHH842 pKa = 5.28VAGTIAASTNTTTGVAGVAPDD863 pKa = 3.71VRR865 pKa = 11.84IMPIRR870 pKa = 11.84VLGEE874 pKa = 4.16GGSGASYY881 pKa = 11.12DD882 pKa = 3.72VAQGILYY889 pKa = 10.41AAGLANDD896 pKa = 4.16SATVPPQAADD906 pKa = 3.78IINLSLGGAYY916 pKa = 10.11VNQASKK922 pKa = 11.11DD923 pKa = 3.86AISAALNAGVIVIAAKK939 pKa = 10.91GNDD942 pKa = 3.93GSDD945 pKa = 3.41APHH948 pKa = 6.48YY949 pKa = 9.75PSDD952 pKa = 3.87YY953 pKa = 10.22EE954 pKa = 4.37GVVSVGALGMGDD966 pKa = 3.35EE967 pKa = 4.1QAYY970 pKa = 8.77YY971 pKa = 11.0SNYY974 pKa = 9.9GPTLDD979 pKa = 4.62LVAPGGDD986 pKa = 3.38SKK988 pKa = 11.19IDD990 pKa = 3.53ADD992 pKa = 4.02NDD994 pKa = 3.7GKK996 pKa = 10.82FDD998 pKa = 3.74STILSTSVDD1007 pKa = 3.38VSSGSRR1013 pKa = 11.84EE1014 pKa = 3.56SGYY1017 pKa = 9.42DD1018 pKa = 3.35HH1019 pKa = 7.3KK1020 pKa = 11.07QGTSMATPHH1029 pKa = 5.58VAGVIALMKK1038 pKa = 10.16SVYY1041 pKa = 10.39SEE1043 pKa = 4.07LTPTLLTALIEE1054 pKa = 4.43NGAITTDD1061 pKa = 3.73LSGDD1065 pKa = 3.5GAAVRR1070 pKa = 11.84NDD1072 pKa = 2.98TYY1074 pKa = 11.53GYY1076 pKa = 11.1GKK1078 pKa = 9.61IDD1080 pKa = 3.47ALKK1083 pKa = 10.55AVQQAQAIAGGATPPVHH1100 pKa = 7.3DD1101 pKa = 4.48EE1102 pKa = 3.69VGIIYY1107 pKa = 10.07EE1108 pKa = 4.08IDD1110 pKa = 3.51NMVLSSSQTISIVKK1124 pKa = 9.22WSGTFTVDD1132 pKa = 5.15SITLNYY1138 pKa = 9.16PWATLTPTAVDD1149 pKa = 3.67GEE1151 pKa = 4.75GFGDD1155 pKa = 3.82YY1156 pKa = 10.66QLDD1159 pKa = 3.31IDD1161 pKa = 4.72RR1162 pKa = 11.84DD1163 pKa = 3.76QFGDD1167 pKa = 3.67GLYY1170 pKa = 10.98LLDD1173 pKa = 3.53ITFVMSNGDD1182 pKa = 3.41QYY1184 pKa = 9.86VTRR1187 pKa = 11.84INVTDD1192 pKa = 3.71RR1193 pKa = 11.84VSTLDD1198 pKa = 3.39EE1199 pKa = 4.34RR1200 pKa = 11.84DD1201 pKa = 3.28VGNVYY1206 pKa = 9.59MVLVDD1211 pKa = 4.29EE1212 pKa = 4.6STLNVVKK1219 pKa = 10.26QIRR1222 pKa = 11.84MNLDD1226 pKa = 2.83VALFEE1231 pKa = 4.67PEE1233 pKa = 4.52VPFSMSNVPDD1243 pKa = 3.74GTYY1246 pKa = 11.05LLYY1249 pKa = 10.88AGTDD1253 pKa = 3.75FDD1255 pKa = 5.3GDD1257 pKa = 3.62RR1258 pKa = 11.84LLEE1261 pKa = 4.48DD1262 pKa = 4.1DD1263 pKa = 4.8EE1264 pKa = 4.89YY1265 pKa = 11.1FGRR1268 pKa = 11.84GFYY1271 pKa = 11.04DD1272 pKa = 3.33HH1273 pKa = 8.0DD1274 pKa = 4.88DD1275 pKa = 4.96DD1276 pKa = 6.06IFTSDD1281 pKa = 3.08ISPIVVNSADD1291 pKa = 3.45IDD1293 pKa = 4.11SVSIYY1298 pKa = 10.68VGSTSADD1305 pKa = 3.28PVGLIDD1311 pKa = 4.31
MM1 pKa = 6.96STNEE5 pKa = 4.0APSASFIVDD14 pKa = 3.39VGTGHH19 pKa = 6.61SPLMVNFDD27 pKa = 4.56ASPSSDD33 pKa = 3.07SDD35 pKa = 3.71GSIAGYY41 pKa = 9.92SWLFGDD47 pKa = 5.12GSTATGQQVSHH58 pKa = 6.98SYY60 pKa = 10.47TNVGNHH66 pKa = 5.5SVTLTVTDD74 pKa = 3.57NEE76 pKa = 4.4GAIDD80 pKa = 3.86NATPQTISVWIANNAPTALFSVDD103 pKa = 3.28TDD105 pKa = 3.64SGEE108 pKa = 4.36GPLTVNFDD116 pKa = 3.52ASASGDD122 pKa = 3.45SDD124 pKa = 3.75GTIVAYY130 pKa = 9.89GWDD133 pKa = 3.87FGDD136 pKa = 3.96GSSGNGQTVAHH147 pKa = 7.09SYY149 pKa = 6.39TTPGVYY155 pKa = 10.2LATLTVRR162 pKa = 11.84DD163 pKa = 4.28DD164 pKa = 3.91DD165 pKa = 4.52AATKK169 pKa = 10.65LSTAHH174 pKa = 7.71IIRR177 pKa = 11.84VLPANQAPTAAFSVDD192 pKa = 3.45TDD194 pKa = 3.51SGTTPLTVNFDD205 pKa = 3.69ASGSTDD211 pKa = 3.11TDD213 pKa = 3.8GSIVSYY219 pKa = 10.76AWDD222 pKa = 4.21FGDD225 pKa = 4.03GDD227 pKa = 4.16SGVGQQVSHH236 pKa = 7.16TYY238 pKa = 10.08TSDD241 pKa = 2.97GSYY244 pKa = 9.36TATLTVTDD252 pKa = 3.73NAGAEE257 pKa = 4.27DD258 pKa = 4.24SDD260 pKa = 4.47SDD262 pKa = 4.45VIDD265 pKa = 4.76SISASHH271 pKa = 6.49YY272 pKa = 6.45PTARR276 pKa = 11.84YY277 pKa = 9.51DD278 pKa = 3.41VDD280 pKa = 3.9VDD282 pKa = 3.97SGPAPLTVSFDD293 pKa = 3.6ASTSSDD299 pKa = 3.65PDD301 pKa = 3.24NDD303 pKa = 3.71IINYY307 pKa = 8.04FWQFGDD313 pKa = 3.55GDD315 pKa = 3.98NASGVTTSHH324 pKa = 7.15TYY326 pKa = 8.4TVPGSYY332 pKa = 10.0DD333 pKa = 3.49VQMLATDD340 pKa = 3.86SASQGDD346 pKa = 4.08VADD349 pKa = 3.55VRR351 pKa = 11.84TITVQGPNTLPTAAFTVDD369 pKa = 3.77VNSGTSPVTVNFDD382 pKa = 3.2ATTSSDD388 pKa = 2.96SDD390 pKa = 3.96GSVVGYY396 pKa = 10.19GWDD399 pKa = 3.7FGDD402 pKa = 4.42GFQGSGASTTHH413 pKa = 6.62TYY415 pKa = 10.2FADD418 pKa = 4.17GTHH421 pKa = 5.75TAILTVTDD429 pKa = 3.95DD430 pKa = 3.53RR431 pKa = 11.84GGQHH435 pKa = 6.09SVSHH439 pKa = 7.35DD440 pKa = 3.09ISVAASGGVTLSGVLGLSGTGTVDD464 pKa = 4.25SDD466 pKa = 4.22VNDD469 pKa = 3.41INADD473 pKa = 3.66YY474 pKa = 10.94QSNNTTATAQLLISPSSLGNVWGFVSKK501 pKa = 9.19TATGIEE507 pKa = 4.44GEE509 pKa = 4.27RR510 pKa = 11.84FTYY513 pKa = 10.52SADD516 pKa = 3.32EE517 pKa = 3.94VDD519 pKa = 3.91IYY521 pKa = 10.88RR522 pKa = 11.84AYY524 pKa = 10.93LPVGQRR530 pKa = 11.84ICLTIKK536 pKa = 10.87NPDD539 pKa = 3.79DD540 pKa = 3.54EE541 pKa = 5.02TGLEE545 pKa = 4.08NANDD549 pKa = 4.13LDD551 pKa = 4.49LYY553 pKa = 10.61LYY555 pKa = 10.91SVDD558 pKa = 4.88DD559 pKa = 4.23PEE561 pKa = 5.91SPVATALGTHH571 pKa = 5.89SSEE574 pKa = 4.3TLHH577 pKa = 6.4VPTGGEE583 pKa = 4.07YY584 pKa = 10.19YY585 pKa = 10.08IVVAAYY591 pKa = 9.97RR592 pKa = 11.84GKK594 pKa = 8.98STYY597 pKa = 10.95KK598 pKa = 10.46LATFMDD604 pKa = 4.55LTSVDD609 pKa = 3.6EE610 pKa = 4.45VQTLRR615 pKa = 11.84LEE617 pKa = 4.53DD618 pKa = 4.31DD619 pKa = 4.44FVPGQVIAKK628 pKa = 9.53LKK630 pKa = 10.54SEE632 pKa = 4.3VVNPPGAGADD642 pKa = 3.58KK643 pKa = 10.71KK644 pKa = 11.11GVAAATAIEE653 pKa = 4.57SVSSRR658 pKa = 11.84GSVSMIRR665 pKa = 11.84GAADD669 pKa = 3.64RR670 pKa = 11.84ASLFSVNDD678 pKa = 3.3MGQNSASGKK687 pKa = 8.53LQSAASSDD695 pKa = 3.65QDD697 pKa = 3.25EE698 pKa = 4.85SEE700 pKa = 4.15YY701 pKa = 11.53SLLDD705 pKa = 3.11GRR707 pKa = 11.84YY708 pKa = 10.32GIGLSQKK715 pKa = 10.26DD716 pKa = 3.26RR717 pKa = 11.84EE718 pKa = 4.34AFEE721 pKa = 4.3TIKK724 pKa = 10.69AIKK727 pKa = 10.03RR728 pKa = 11.84LRR730 pKa = 11.84QDD732 pKa = 2.64SRR734 pKa = 11.84IDD736 pKa = 3.56YY737 pKa = 10.83AEE739 pKa = 4.12PNYY742 pKa = 8.52IQKK745 pKa = 9.74MSLVPDD751 pKa = 4.34DD752 pKa = 4.45LHH754 pKa = 8.86YY755 pKa = 10.17EE756 pKa = 4.35QQWHH760 pKa = 5.72YY761 pKa = 10.35PAINLPDD768 pKa = 3.17TWDD771 pKa = 3.26ITTGDD776 pKa = 3.37ASVIVAVIDD785 pKa = 3.52SGVFMAHH792 pKa = 6.89EE793 pKa = 4.67DD794 pKa = 3.45LSGNLLSTGYY804 pKa = 10.77DD805 pKa = 3.88FIVSTSMSNDD815 pKa = 2.87GDD817 pKa = 4.88GIDD820 pKa = 5.14PDD822 pKa = 5.2PDD824 pKa = 4.6PDD826 pKa = 4.13DD827 pKa = 5.11PGDD830 pKa = 3.6IVISYY835 pKa = 7.69SVWHH839 pKa = 5.97GTHH842 pKa = 5.28VAGTIAASTNTTTGVAGVAPDD863 pKa = 3.71VRR865 pKa = 11.84IMPIRR870 pKa = 11.84VLGEE874 pKa = 4.16GGSGASYY881 pKa = 11.12DD882 pKa = 3.72VAQGILYY889 pKa = 10.41AAGLANDD896 pKa = 4.16SATVPPQAADD906 pKa = 3.78IINLSLGGAYY916 pKa = 10.11VNQASKK922 pKa = 11.11DD923 pKa = 3.86AISAALNAGVIVIAAKK939 pKa = 10.91GNDD942 pKa = 3.93GSDD945 pKa = 3.41APHH948 pKa = 6.48YY949 pKa = 9.75PSDD952 pKa = 3.87YY953 pKa = 10.22EE954 pKa = 4.37GVVSVGALGMGDD966 pKa = 3.35EE967 pKa = 4.1QAYY970 pKa = 8.77YY971 pKa = 11.0SNYY974 pKa = 9.9GPTLDD979 pKa = 4.62LVAPGGDD986 pKa = 3.38SKK988 pKa = 11.19IDD990 pKa = 3.53ADD992 pKa = 4.02NDD994 pKa = 3.7GKK996 pKa = 10.82FDD998 pKa = 3.74STILSTSVDD1007 pKa = 3.38VSSGSRR1013 pKa = 11.84EE1014 pKa = 3.56SGYY1017 pKa = 9.42DD1018 pKa = 3.35HH1019 pKa = 7.3KK1020 pKa = 11.07QGTSMATPHH1029 pKa = 5.58VAGVIALMKK1038 pKa = 10.16SVYY1041 pKa = 10.39SEE1043 pKa = 4.07LTPTLLTALIEE1054 pKa = 4.43NGAITTDD1061 pKa = 3.73LSGDD1065 pKa = 3.5GAAVRR1070 pKa = 11.84NDD1072 pKa = 2.98TYY1074 pKa = 11.53GYY1076 pKa = 11.1GKK1078 pKa = 9.61IDD1080 pKa = 3.47ALKK1083 pKa = 10.55AVQQAQAIAGGATPPVHH1100 pKa = 7.3DD1101 pKa = 4.48EE1102 pKa = 3.69VGIIYY1107 pKa = 10.07EE1108 pKa = 4.08IDD1110 pKa = 3.51NMVLSSSQTISIVKK1124 pKa = 9.22WSGTFTVDD1132 pKa = 5.15SITLNYY1138 pKa = 9.16PWATLTPTAVDD1149 pKa = 3.67GEE1151 pKa = 4.75GFGDD1155 pKa = 3.82YY1156 pKa = 10.66QLDD1159 pKa = 3.31IDD1161 pKa = 4.72RR1162 pKa = 11.84DD1163 pKa = 3.76QFGDD1167 pKa = 3.67GLYY1170 pKa = 10.98LLDD1173 pKa = 3.53ITFVMSNGDD1182 pKa = 3.41QYY1184 pKa = 9.86VTRR1187 pKa = 11.84INVTDD1192 pKa = 3.71RR1193 pKa = 11.84VSTLDD1198 pKa = 3.39EE1199 pKa = 4.34RR1200 pKa = 11.84DD1201 pKa = 3.28VGNVYY1206 pKa = 9.59MVLVDD1211 pKa = 4.29EE1212 pKa = 4.6STLNVVKK1219 pKa = 10.26QIRR1222 pKa = 11.84MNLDD1226 pKa = 2.83VALFEE1231 pKa = 4.67PEE1233 pKa = 4.52VPFSMSNVPDD1243 pKa = 3.74GTYY1246 pKa = 11.05LLYY1249 pKa = 10.88AGTDD1253 pKa = 3.75FDD1255 pKa = 5.3GDD1257 pKa = 3.62RR1258 pKa = 11.84LLEE1261 pKa = 4.48DD1262 pKa = 4.1DD1263 pKa = 4.8EE1264 pKa = 4.89YY1265 pKa = 11.1FGRR1268 pKa = 11.84GFYY1271 pKa = 11.04DD1272 pKa = 3.33HH1273 pKa = 8.0DD1274 pKa = 4.88DD1275 pKa = 4.96DD1276 pKa = 6.06IFTSDD1281 pKa = 3.08ISPIVVNSADD1291 pKa = 3.45IDD1293 pKa = 4.11SVSIYY1298 pKa = 10.68VGSTSADD1305 pKa = 3.28PVGLIDD1311 pKa = 4.31
Molecular weight: 137.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1T2L4W3|A0A1T2L4W3_9GAMM Phosphoribosylformylglycinamidine synthase OS=Solemya pervernicosa gill symbiont OX=642797 GN=purL PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 9.97RR12 pKa = 11.84KK13 pKa = 8.91RR14 pKa = 11.84VHH16 pKa = 6.24GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.06NGRR28 pKa = 11.84KK29 pKa = 9.38VISARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.3GRR39 pKa = 11.84ARR41 pKa = 11.84LCAA44 pKa = 3.92
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 9.97RR12 pKa = 11.84KK13 pKa = 8.91RR14 pKa = 11.84VHH16 pKa = 6.24GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.06NGRR28 pKa = 11.84KK29 pKa = 9.38VISARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.3GRR39 pKa = 11.84ARR41 pKa = 11.84LCAA44 pKa = 3.92
Molecular weight: 5.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1026818 |
30 |
4114 |
317.3 |
35.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.193 ± 0.048 | 0.958 ± 0.022 |
5.86 ± 0.039 | 7.076 ± 0.046 |
3.581 ± 0.026 | 7.594 ± 0.088 |
2.382 ± 0.025 | 6.021 ± 0.034 |
4.155 ± 0.048 | 10.506 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.61 ± 0.026 | 3.567 ± 0.032 |
4.114 ± 0.033 | 3.863 ± 0.027 |
5.998 ± 0.049 | 6.538 ± 0.053 |
5.119 ± 0.039 | 6.932 ± 0.04 |
1.175 ± 0.018 | 2.76 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
