
Lynx canadensis faeces associated genomovirus CL4 71
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; unclassified Genomoviridae
Average proteome isoelectric point is 7.25
Get precalculated fractions of proteins
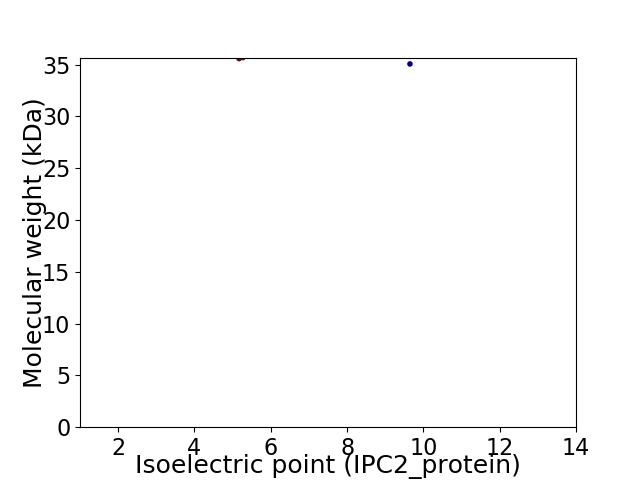
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5CK53|A0A2Z5CK53_9VIRU Replication-associated protein OS=Lynx canadensis faeces associated genomovirus CL4 71 OX=2219131 PE=3 SV=1
MM1 pKa = 7.89PSFHH5 pKa = 6.86LKK7 pKa = 8.75NRR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 10.69VLFTYY16 pKa = 10.1AQAGSDD22 pKa = 3.66FDD24 pKa = 3.31HH25 pKa = 6.81WAVVDD30 pKa = 4.09LLGGLGAEE38 pKa = 4.56CIIGRR43 pKa = 11.84EE44 pKa = 4.06SHH46 pKa = 6.95ADD48 pKa = 3.3GGVHH52 pKa = 5.47FHH54 pKa = 6.24VFADD58 pKa = 4.63FGRR61 pKa = 11.84LFSTRR66 pKa = 11.84KK67 pKa = 8.13TDD69 pKa = 3.59LFDD72 pKa = 4.25VGGKK76 pKa = 9.29HH77 pKa = 6.83PNIQPIGRR85 pKa = 11.84TPAKK89 pKa = 10.3AYY91 pKa = 10.39DD92 pKa = 3.93YY93 pKa = 10.86ACKK96 pKa = 10.72DD97 pKa = 3.23GDD99 pKa = 4.09VVAGGLGRR107 pKa = 11.84PGGDD111 pKa = 3.43ADD113 pKa = 4.94FDD115 pKa = 4.67PDD117 pKa = 4.67SFWDD121 pKa = 3.7SATHH125 pKa = 6.59CGSADD130 pKa = 3.65EE131 pKa = 4.98FLHH134 pKa = 7.02FCDD137 pKa = 4.14QLAPRR142 pKa = 11.84DD143 pKa = 4.09LIRR146 pKa = 11.84GFPAFRR152 pKa = 11.84SYY154 pKa = 11.85SNWKK158 pKa = 8.47WNDD161 pKa = 2.84GALVYY166 pKa = 9.43NQPDD170 pKa = 3.52GVEE173 pKa = 4.29FDD175 pKa = 3.38TSAAEE180 pKa = 4.92GIDD183 pKa = 3.48EE184 pKa = 4.26RR185 pKa = 11.84RR186 pKa = 11.84SLVLFGPYY194 pKa = 10.43GCGKK198 pKa = 5.65TTWARR203 pKa = 11.84SLGEE207 pKa = 4.02HH208 pKa = 6.89IYY210 pKa = 10.68FGSQWSGKK218 pKa = 7.76VAFQGLEE225 pKa = 3.77KK226 pKa = 10.81AEE228 pKa = 3.85YY229 pKa = 10.37AIFDD233 pKa = 3.71DD234 pKa = 3.95WKK236 pKa = 10.84GGLKK240 pKa = 9.14MLPGYY245 pKa = 10.21KK246 pKa = 9.65DD247 pKa = 2.9WLGAQWHH254 pKa = 5.04VSVRR258 pKa = 11.84QLHH261 pKa = 6.14HH262 pKa = 7.14DD263 pKa = 3.37ARR265 pKa = 11.84LEE267 pKa = 3.93EE268 pKa = 4.12WGRR271 pKa = 11.84PCIWLCNTDD280 pKa = 3.88PRR282 pKa = 11.84VMTHH286 pKa = 6.19VNDD289 pKa = 3.84DD290 pKa = 4.22TDD292 pKa = 3.91WTWMDD297 pKa = 3.94RR298 pKa = 11.84ACIFVEE304 pKa = 4.36VTGVLATFRR313 pKa = 11.84ASTEE317 pKa = 3.78
MM1 pKa = 7.89PSFHH5 pKa = 6.86LKK7 pKa = 8.75NRR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 10.69VLFTYY16 pKa = 10.1AQAGSDD22 pKa = 3.66FDD24 pKa = 3.31HH25 pKa = 6.81WAVVDD30 pKa = 4.09LLGGLGAEE38 pKa = 4.56CIIGRR43 pKa = 11.84EE44 pKa = 4.06SHH46 pKa = 6.95ADD48 pKa = 3.3GGVHH52 pKa = 5.47FHH54 pKa = 6.24VFADD58 pKa = 4.63FGRR61 pKa = 11.84LFSTRR66 pKa = 11.84KK67 pKa = 8.13TDD69 pKa = 3.59LFDD72 pKa = 4.25VGGKK76 pKa = 9.29HH77 pKa = 6.83PNIQPIGRR85 pKa = 11.84TPAKK89 pKa = 10.3AYY91 pKa = 10.39DD92 pKa = 3.93YY93 pKa = 10.86ACKK96 pKa = 10.72DD97 pKa = 3.23GDD99 pKa = 4.09VVAGGLGRR107 pKa = 11.84PGGDD111 pKa = 3.43ADD113 pKa = 4.94FDD115 pKa = 4.67PDD117 pKa = 4.67SFWDD121 pKa = 3.7SATHH125 pKa = 6.59CGSADD130 pKa = 3.65EE131 pKa = 4.98FLHH134 pKa = 7.02FCDD137 pKa = 4.14QLAPRR142 pKa = 11.84DD143 pKa = 4.09LIRR146 pKa = 11.84GFPAFRR152 pKa = 11.84SYY154 pKa = 11.85SNWKK158 pKa = 8.47WNDD161 pKa = 2.84GALVYY166 pKa = 9.43NQPDD170 pKa = 3.52GVEE173 pKa = 4.29FDD175 pKa = 3.38TSAAEE180 pKa = 4.92GIDD183 pKa = 3.48EE184 pKa = 4.26RR185 pKa = 11.84RR186 pKa = 11.84SLVLFGPYY194 pKa = 10.43GCGKK198 pKa = 5.65TTWARR203 pKa = 11.84SLGEE207 pKa = 4.02HH208 pKa = 6.89IYY210 pKa = 10.68FGSQWSGKK218 pKa = 7.76VAFQGLEE225 pKa = 3.77KK226 pKa = 10.81AEE228 pKa = 3.85YY229 pKa = 10.37AIFDD233 pKa = 3.71DD234 pKa = 3.95WKK236 pKa = 10.84GGLKK240 pKa = 9.14MLPGYY245 pKa = 10.21KK246 pKa = 9.65DD247 pKa = 2.9WLGAQWHH254 pKa = 5.04VSVRR258 pKa = 11.84QLHH261 pKa = 6.14HH262 pKa = 7.14DD263 pKa = 3.37ARR265 pKa = 11.84LEE267 pKa = 3.93EE268 pKa = 4.12WGRR271 pKa = 11.84PCIWLCNTDD280 pKa = 3.88PRR282 pKa = 11.84VMTHH286 pKa = 6.19VNDD289 pKa = 3.84DD290 pKa = 4.22TDD292 pKa = 3.91WTWMDD297 pKa = 3.94RR298 pKa = 11.84ACIFVEE304 pKa = 4.36VTGVLATFRR313 pKa = 11.84ASTEE317 pKa = 3.78
Molecular weight: 35.58 kDa
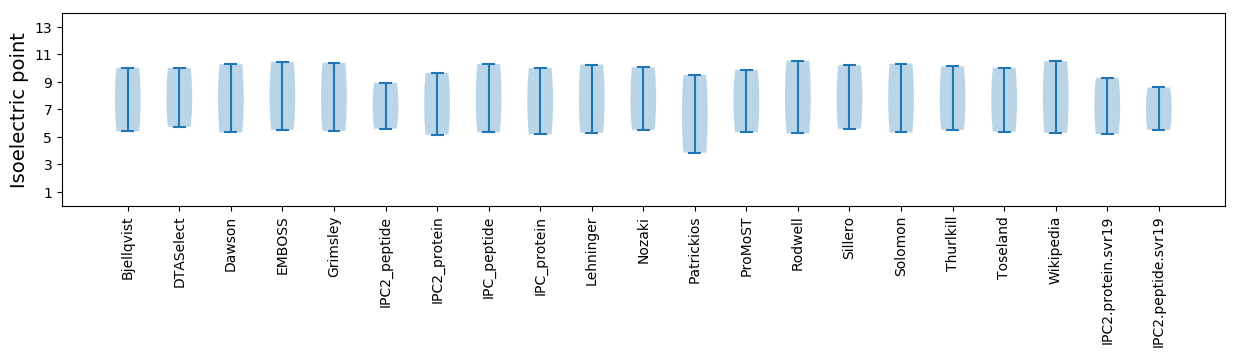
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CK53|A0A2Z5CK53_9VIRU Replication-associated protein OS=Lynx canadensis faeces associated genomovirus CL4 71 OX=2219131 PE=3 SV=1
MM1 pKa = 7.82AYY3 pKa = 10.14ARR5 pKa = 11.84RR6 pKa = 11.84PRR8 pKa = 11.84RR9 pKa = 11.84PAYY12 pKa = 9.27RR13 pKa = 11.84KK14 pKa = 9.61KK15 pKa = 10.2GARR18 pKa = 11.84STRR21 pKa = 11.84RR22 pKa = 11.84YY23 pKa = 9.83SKK25 pKa = 10.25RR26 pKa = 11.84SYY28 pKa = 10.02PRR30 pKa = 11.84SSYY33 pKa = 10.09VKK35 pKa = 9.18RR36 pKa = 11.84RR37 pKa = 11.84PYY39 pKa = 10.0RR40 pKa = 11.84KK41 pKa = 9.87RR42 pKa = 11.84MTKK45 pKa = 10.34KK46 pKa = 10.35SILNVTSRR54 pKa = 11.84KK55 pKa = 9.78KK56 pKa = 10.11RR57 pKa = 11.84DD58 pKa = 2.95TMAYY62 pKa = 6.99YY63 pKa = 10.34TNSTAASQTGGTTYY77 pKa = 10.72VQDD80 pKa = 3.58AAVVTGGTSVPACFLWCATGRR101 pKa = 11.84DD102 pKa = 4.19YY103 pKa = 8.2TTSNAGTNGQIVDD116 pKa = 3.52AASRR120 pKa = 11.84TSQSCYY126 pKa = 8.3MRR128 pKa = 11.84GLKK131 pKa = 9.53EE132 pKa = 3.77ALEE135 pKa = 4.29VQVADD140 pKa = 3.88GVPWQWRR147 pKa = 11.84RR148 pKa = 11.84ICFTAKK154 pKa = 10.43GLQTLFAATTGFSLVIEE171 pKa = 4.8ASTGYY176 pKa = 10.19QRR178 pKa = 11.84VLNQIPGDD186 pKa = 3.97SATATSLRR194 pKa = 11.84AVIEE198 pKa = 4.14TVLFKK203 pKa = 10.78GVYY206 pKa = 9.22LRR208 pKa = 11.84DD209 pKa = 3.7WIDD212 pKa = 3.43PMTASTDD219 pKa = 3.35NDD221 pKa = 3.79RR222 pKa = 11.84LTIKK226 pKa = 10.24YY227 pKa = 9.92DD228 pKa = 3.27KK229 pKa = 10.2TITIASGNEE238 pKa = 3.72DD239 pKa = 3.41GCIRR243 pKa = 11.84NYY245 pKa = 10.33KK246 pKa = 9.06RR247 pKa = 11.84WHH249 pKa = 6.12GMNKK253 pKa = 8.33TLVYY257 pKa = 10.85NDD259 pKa = 4.72DD260 pKa = 3.63EE261 pKa = 5.35SGGTKK266 pKa = 10.0VPAFYY271 pKa = 9.7STLGKK276 pKa = 10.56AGMGDD281 pKa = 4.51FYY283 pKa = 11.06IMDD286 pKa = 4.19YY287 pKa = 10.35FRR289 pKa = 11.84PRR291 pKa = 11.84AGSTSTNQLRR301 pKa = 11.84VSASSTLYY309 pKa = 9.07WHH311 pKa = 7.06EE312 pKa = 4.1KK313 pKa = 9.32
MM1 pKa = 7.82AYY3 pKa = 10.14ARR5 pKa = 11.84RR6 pKa = 11.84PRR8 pKa = 11.84RR9 pKa = 11.84PAYY12 pKa = 9.27RR13 pKa = 11.84KK14 pKa = 9.61KK15 pKa = 10.2GARR18 pKa = 11.84STRR21 pKa = 11.84RR22 pKa = 11.84YY23 pKa = 9.83SKK25 pKa = 10.25RR26 pKa = 11.84SYY28 pKa = 10.02PRR30 pKa = 11.84SSYY33 pKa = 10.09VKK35 pKa = 9.18RR36 pKa = 11.84RR37 pKa = 11.84PYY39 pKa = 10.0RR40 pKa = 11.84KK41 pKa = 9.87RR42 pKa = 11.84MTKK45 pKa = 10.34KK46 pKa = 10.35SILNVTSRR54 pKa = 11.84KK55 pKa = 9.78KK56 pKa = 10.11RR57 pKa = 11.84DD58 pKa = 2.95TMAYY62 pKa = 6.99YY63 pKa = 10.34TNSTAASQTGGTTYY77 pKa = 10.72VQDD80 pKa = 3.58AAVVTGGTSVPACFLWCATGRR101 pKa = 11.84DD102 pKa = 4.19YY103 pKa = 8.2TTSNAGTNGQIVDD116 pKa = 3.52AASRR120 pKa = 11.84TSQSCYY126 pKa = 8.3MRR128 pKa = 11.84GLKK131 pKa = 9.53EE132 pKa = 3.77ALEE135 pKa = 4.29VQVADD140 pKa = 3.88GVPWQWRR147 pKa = 11.84RR148 pKa = 11.84ICFTAKK154 pKa = 10.43GLQTLFAATTGFSLVIEE171 pKa = 4.8ASTGYY176 pKa = 10.19QRR178 pKa = 11.84VLNQIPGDD186 pKa = 3.97SATATSLRR194 pKa = 11.84AVIEE198 pKa = 4.14TVLFKK203 pKa = 10.78GVYY206 pKa = 9.22LRR208 pKa = 11.84DD209 pKa = 3.7WIDD212 pKa = 3.43PMTASTDD219 pKa = 3.35NDD221 pKa = 3.79RR222 pKa = 11.84LTIKK226 pKa = 10.24YY227 pKa = 9.92DD228 pKa = 3.27KK229 pKa = 10.2TITIASGNEE238 pKa = 3.72DD239 pKa = 3.41GCIRR243 pKa = 11.84NYY245 pKa = 10.33KK246 pKa = 9.06RR247 pKa = 11.84WHH249 pKa = 6.12GMNKK253 pKa = 8.33TLVYY257 pKa = 10.85NDD259 pKa = 4.72DD260 pKa = 3.63EE261 pKa = 5.35SGGTKK266 pKa = 10.0VPAFYY271 pKa = 9.7STLGKK276 pKa = 10.56AGMGDD281 pKa = 4.51FYY283 pKa = 11.06IMDD286 pKa = 4.19YY287 pKa = 10.35FRR289 pKa = 11.84PRR291 pKa = 11.84AGSTSTNQLRR301 pKa = 11.84VSASSTLYY309 pKa = 9.07WHH311 pKa = 7.06EE312 pKa = 4.1KK313 pKa = 9.32
Molecular weight: 35.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
630 |
313 |
317 |
315.0 |
35.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.889 ± 0.253 | 2.063 ± 0.313 |
7.46 ± 1.577 | 3.175 ± 0.63 |
4.603 ± 1.375 | 9.365 ± 1.14 |
2.381 ± 1.17 | 3.492 ± 0.23 |
4.921 ± 0.772 | 6.19 ± 0.724 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.905 ± 0.437 | 2.857 ± 0.441 |
3.651 ± 0.306 | 2.857 ± 0.227 |
7.778 ± 1.214 | 6.825 ± 1.209 |
7.937 ± 2.18 | 5.873 ± 0.082 |
3.016 ± 0.738 | 4.762 ± 1.093 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
