
Human papillomavirus 22
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Betapapillomavirus; Betapapillomavirus 2
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
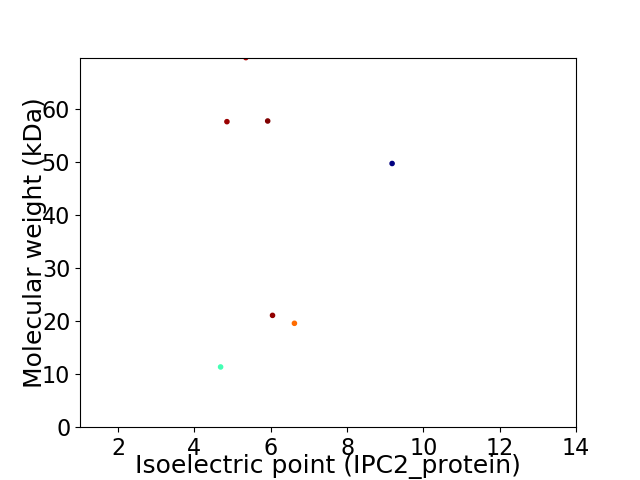
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P50788|VL1_HPV22 Major capsid protein L1 OS=Human papillomavirus 22 OX=37954 GN=L1 PE=3 SV=1
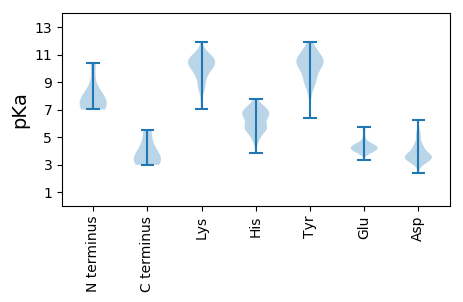
MM1 pKa = 7.54IGKK4 pKa = 8.5QATLCDD10 pKa = 3.55IVLEE14 pKa = 4.14EE15 pKa = 4.64LVLPIDD21 pKa = 3.84LHH23 pKa = 6.32CHH25 pKa = 5.61EE26 pKa = 5.67EE27 pKa = 4.29LPEE30 pKa = 4.22LPEE33 pKa = 4.02EE34 pKa = 4.36LEE36 pKa = 3.99EE37 pKa = 4.46SVVEE41 pKa = 4.17EE42 pKa = 4.18EE43 pKa = 5.43PEE45 pKa = 3.87YY46 pKa = 10.33TPYY49 pKa = 10.77KK50 pKa = 9.65IVVYY54 pKa = 10.65CGGCDD59 pKa = 3.31TKK61 pKa = 11.31LKK63 pKa = 10.71LYY65 pKa = 10.38ILATLSGIRR74 pKa = 11.84DD75 pKa = 3.89FQTSLLGPVKK85 pKa = 10.12LLCPTCRR92 pKa = 11.84EE93 pKa = 4.03EE94 pKa = 4.14IRR96 pKa = 11.84NGRR99 pKa = 11.84RR100 pKa = 3.02
MM1 pKa = 7.54IGKK4 pKa = 8.5QATLCDD10 pKa = 3.55IVLEE14 pKa = 4.14EE15 pKa = 4.64LVLPIDD21 pKa = 3.84LHH23 pKa = 6.32CHH25 pKa = 5.61EE26 pKa = 5.67EE27 pKa = 4.29LPEE30 pKa = 4.22LPEE33 pKa = 4.02EE34 pKa = 4.36LEE36 pKa = 3.99EE37 pKa = 4.46SVVEE41 pKa = 4.17EE42 pKa = 4.18EE43 pKa = 5.43PEE45 pKa = 3.87YY46 pKa = 10.33TPYY49 pKa = 10.77KK50 pKa = 9.65IVVYY54 pKa = 10.65CGGCDD59 pKa = 3.31TKK61 pKa = 11.31LKK63 pKa = 10.71LYY65 pKa = 10.38ILATLSGIRR74 pKa = 11.84DD75 pKa = 3.89FQTSLLGPVKK85 pKa = 10.12LLCPTCRR92 pKa = 11.84EE93 pKa = 4.03EE94 pKa = 4.14IRR96 pKa = 11.84NGRR99 pKa = 11.84RR100 pKa = 3.02
Molecular weight: 11.36 kDa
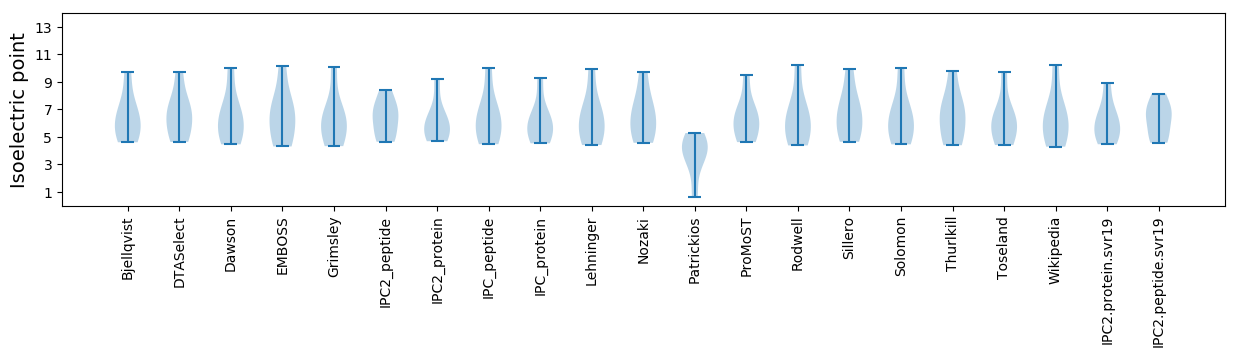
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P50775|VE6_HPV22 Protein E6 OS=Human papillomavirus 22 OX=37954 GN=E6 PE=3 SV=1
MM1 pKa = 7.65EE2 pKa = 5.45KK3 pKa = 10.47LSEE6 pKa = 4.19RR7 pKa = 11.84FSALQEE13 pKa = 4.13KK14 pKa = 10.62LMDD17 pKa = 4.3LYY19 pKa = 11.28EE20 pKa = 5.14SGVEE24 pKa = 3.96DD25 pKa = 5.76LEE27 pKa = 4.52TQIQHH32 pKa = 5.68WKK34 pKa = 10.31LLRR37 pKa = 11.84QEE39 pKa = 4.1QVLFYY44 pKa = 9.06YY45 pKa = 10.58ARR47 pKa = 11.84RR48 pKa = 11.84HH49 pKa = 5.52GILRR53 pKa = 11.84LGYY56 pKa = 10.15QPVPTLATSEE66 pKa = 4.56SKK68 pKa = 11.11AKK70 pKa = 10.43DD71 pKa = 3.81AIAMGLLLEE80 pKa = 4.66SLQKK84 pKa = 10.32SQYY87 pKa = 10.93AEE89 pKa = 4.19EE90 pKa = 4.41PWTLVEE96 pKa = 4.35TSLEE100 pKa = 4.49TVKK103 pKa = 10.9SPPADD108 pKa = 3.66CFKK111 pKa = 10.81KK112 pKa = 10.31GPKK115 pKa = 9.57SVEE118 pKa = 4.01VYY120 pKa = 10.71FDD122 pKa = 3.98GDD124 pKa = 3.74PEE126 pKa = 4.3NVMSYY131 pKa = 7.4TVWSYY136 pKa = 11.33IYY138 pKa = 10.26YY139 pKa = 8.54QTDD142 pKa = 3.26DD143 pKa = 3.88EE144 pKa = 4.49SWEE147 pKa = 4.15KK148 pKa = 11.3VEE150 pKa = 4.39GHH152 pKa = 5.67VDD154 pKa = 3.38YY155 pKa = 11.04TGAYY159 pKa = 8.88YY160 pKa = 9.92IEE162 pKa = 4.73GTFKK166 pKa = 10.02TYY168 pKa = 10.46YY169 pKa = 10.05IKK171 pKa = 10.91FEE173 pKa = 4.25TDD175 pKa = 2.4AKK177 pKa = 10.55RR178 pKa = 11.84YY179 pKa = 6.43GTTGHH184 pKa = 6.66WEE186 pKa = 3.74VHH188 pKa = 4.76VNKK191 pKa = 9.36DD192 pKa = 3.53TVFTPVTSSTPPVGVASQNSAPEE215 pKa = 4.1PASTSDD221 pKa = 3.09SPQRR225 pKa = 11.84SSQVTHH231 pKa = 6.93RR232 pKa = 11.84YY233 pKa = 8.31GRR235 pKa = 11.84KK236 pKa = 9.38ASSPTITTIRR246 pKa = 11.84RR247 pKa = 11.84QKK249 pKa = 9.53RR250 pKa = 11.84RR251 pKa = 11.84EE252 pKa = 3.86RR253 pKa = 11.84QRR255 pKa = 11.84QEE257 pKa = 3.51TPTRR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84KK264 pKa = 7.37TRR266 pKa = 11.84SRR268 pKa = 11.84SRR270 pKa = 11.84STEE273 pKa = 3.58QRR275 pKa = 11.84GGRR278 pKa = 11.84ATRR281 pKa = 11.84RR282 pKa = 11.84SLSRR286 pKa = 11.84EE287 pKa = 3.82SAEE290 pKa = 3.94SPRR293 pKa = 11.84RR294 pKa = 11.84GGRR297 pKa = 11.84GGGGPLTRR305 pKa = 11.84SRR307 pKa = 11.84SRR309 pKa = 11.84SRR311 pKa = 11.84SRR313 pKa = 11.84TRR315 pKa = 11.84EE316 pKa = 3.95SVDD319 pKa = 3.22GGGVAPDD326 pKa = 3.43EE327 pKa = 4.5VGATLRR333 pKa = 11.84SIGRR337 pKa = 11.84QHH339 pKa = 7.26SGRR342 pKa = 11.84LAQLLDD348 pKa = 3.62AAKK351 pKa = 9.99DD352 pKa = 3.57PPVILLRR359 pKa = 11.84GAANTLKK366 pKa = 10.45CYY368 pKa = 10.33RR369 pKa = 11.84YY370 pKa = 10.07RR371 pKa = 11.84FRR373 pKa = 11.84KK374 pKa = 8.73KK375 pKa = 9.77HH376 pKa = 5.68AGSFQFISTTWSWVGGHH393 pKa = 4.66TTDD396 pKa = 4.03RR397 pKa = 11.84IGRR400 pKa = 11.84SRR402 pKa = 11.84ILISFHH408 pKa = 5.67TDD410 pKa = 3.06RR411 pKa = 11.84EE412 pKa = 4.27RR413 pKa = 11.84EE414 pKa = 3.92KK415 pKa = 11.0CLQQMKK421 pKa = 10.49LPLGVEE427 pKa = 4.08WSYY430 pKa = 11.91GQFDD434 pKa = 4.19DD435 pKa = 5.18LL436 pKa = 5.54
MM1 pKa = 7.65EE2 pKa = 5.45KK3 pKa = 10.47LSEE6 pKa = 4.19RR7 pKa = 11.84FSALQEE13 pKa = 4.13KK14 pKa = 10.62LMDD17 pKa = 4.3LYY19 pKa = 11.28EE20 pKa = 5.14SGVEE24 pKa = 3.96DD25 pKa = 5.76LEE27 pKa = 4.52TQIQHH32 pKa = 5.68WKK34 pKa = 10.31LLRR37 pKa = 11.84QEE39 pKa = 4.1QVLFYY44 pKa = 9.06YY45 pKa = 10.58ARR47 pKa = 11.84RR48 pKa = 11.84HH49 pKa = 5.52GILRR53 pKa = 11.84LGYY56 pKa = 10.15QPVPTLATSEE66 pKa = 4.56SKK68 pKa = 11.11AKK70 pKa = 10.43DD71 pKa = 3.81AIAMGLLLEE80 pKa = 4.66SLQKK84 pKa = 10.32SQYY87 pKa = 10.93AEE89 pKa = 4.19EE90 pKa = 4.41PWTLVEE96 pKa = 4.35TSLEE100 pKa = 4.49TVKK103 pKa = 10.9SPPADD108 pKa = 3.66CFKK111 pKa = 10.81KK112 pKa = 10.31GPKK115 pKa = 9.57SVEE118 pKa = 4.01VYY120 pKa = 10.71FDD122 pKa = 3.98GDD124 pKa = 3.74PEE126 pKa = 4.3NVMSYY131 pKa = 7.4TVWSYY136 pKa = 11.33IYY138 pKa = 10.26YY139 pKa = 8.54QTDD142 pKa = 3.26DD143 pKa = 3.88EE144 pKa = 4.49SWEE147 pKa = 4.15KK148 pKa = 11.3VEE150 pKa = 4.39GHH152 pKa = 5.67VDD154 pKa = 3.38YY155 pKa = 11.04TGAYY159 pKa = 8.88YY160 pKa = 9.92IEE162 pKa = 4.73GTFKK166 pKa = 10.02TYY168 pKa = 10.46YY169 pKa = 10.05IKK171 pKa = 10.91FEE173 pKa = 4.25TDD175 pKa = 2.4AKK177 pKa = 10.55RR178 pKa = 11.84YY179 pKa = 6.43GTTGHH184 pKa = 6.66WEE186 pKa = 3.74VHH188 pKa = 4.76VNKK191 pKa = 9.36DD192 pKa = 3.53TVFTPVTSSTPPVGVASQNSAPEE215 pKa = 4.1PASTSDD221 pKa = 3.09SPQRR225 pKa = 11.84SSQVTHH231 pKa = 6.93RR232 pKa = 11.84YY233 pKa = 8.31GRR235 pKa = 11.84KK236 pKa = 9.38ASSPTITTIRR246 pKa = 11.84RR247 pKa = 11.84QKK249 pKa = 9.53RR250 pKa = 11.84RR251 pKa = 11.84EE252 pKa = 3.86RR253 pKa = 11.84QRR255 pKa = 11.84QEE257 pKa = 3.51TPTRR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84KK264 pKa = 7.37TRR266 pKa = 11.84SRR268 pKa = 11.84SRR270 pKa = 11.84STEE273 pKa = 3.58QRR275 pKa = 11.84GGRR278 pKa = 11.84ATRR281 pKa = 11.84RR282 pKa = 11.84SLSRR286 pKa = 11.84EE287 pKa = 3.82SAEE290 pKa = 3.94SPRR293 pKa = 11.84RR294 pKa = 11.84GGRR297 pKa = 11.84GGGGPLTRR305 pKa = 11.84SRR307 pKa = 11.84SRR309 pKa = 11.84SRR311 pKa = 11.84SRR313 pKa = 11.84TRR315 pKa = 11.84EE316 pKa = 3.95SVDD319 pKa = 3.22GGGVAPDD326 pKa = 3.43EE327 pKa = 4.5VGATLRR333 pKa = 11.84SIGRR337 pKa = 11.84QHH339 pKa = 7.26SGRR342 pKa = 11.84LAQLLDD348 pKa = 3.62AAKK351 pKa = 9.99DD352 pKa = 3.57PPVILLRR359 pKa = 11.84GAANTLKK366 pKa = 10.45CYY368 pKa = 10.33RR369 pKa = 11.84YY370 pKa = 10.07RR371 pKa = 11.84FRR373 pKa = 11.84KK374 pKa = 8.73KK375 pKa = 9.77HH376 pKa = 5.68AGSFQFISTTWSWVGGHH393 pKa = 4.66TTDD396 pKa = 4.03RR397 pKa = 11.84IGRR400 pKa = 11.84SRR402 pKa = 11.84ILISFHH408 pKa = 5.67TDD410 pKa = 3.06RR411 pKa = 11.84EE412 pKa = 4.27RR413 pKa = 11.84EE414 pKa = 3.92KK415 pKa = 11.0CLQQMKK421 pKa = 10.49LPLGVEE427 pKa = 4.08WSYY430 pKa = 11.91GQFDD434 pKa = 4.19DD435 pKa = 5.18LL436 pKa = 5.54
Molecular weight: 49.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2536 |
100 |
608 |
362.3 |
40.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.323 ± 0.456 | 2.169 ± 0.653 |
6.23 ± 0.38 | 6.822 ± 0.592 |
4.101 ± 0.511 | 6.388 ± 0.617 |
2.248 ± 0.228 | 4.771 ± 0.756 |
4.968 ± 0.707 | 8.872 ± 0.717 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.696 ± 0.53 | 3.352 ± 0.694 |
6.467 ± 1.15 | 5.047 ± 0.691 |
6.782 ± 0.992 | 7.571 ± 0.649 |
6.23 ± 0.792 | 6.191 ± 0.802 |
1.222 ± 0.336 | 3.549 ± 0.426 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
