
Fig cryptic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Deltapartitivirus
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
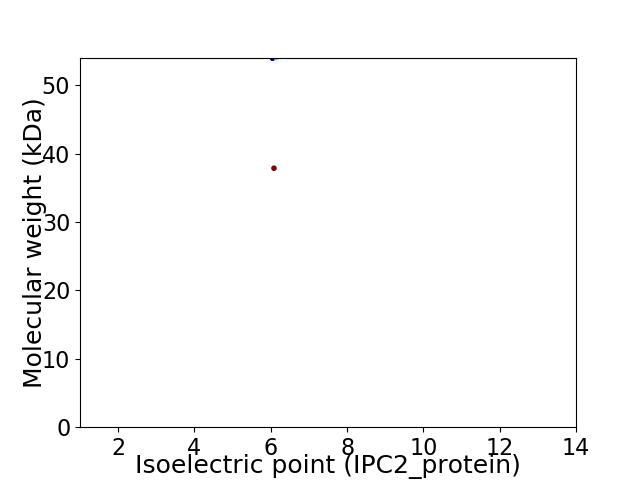
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|F4MST4|F4MST4_9VIRU Putative capsid protein OS=Fig cryptic virus OX=882768 GN=CP PE=4 SV=1
MM1 pKa = 7.41EE2 pKa = 6.01AGLIEE7 pKa = 4.58IGNIPEE13 pKa = 3.69RR14 pKa = 11.84HH15 pKa = 6.05LRR17 pKa = 11.84DD18 pKa = 3.31EE19 pKa = 4.91FIILVDD25 pKa = 3.62QPAYY29 pKa = 10.87DD30 pKa = 3.58SVRR33 pKa = 11.84RR34 pKa = 11.84NAPQADD40 pKa = 3.77MQEE43 pKa = 3.77IDD45 pKa = 2.52GWARR49 pKa = 11.84SFYY52 pKa = 9.43TVEE55 pKa = 4.96GIMASIMQFSKK66 pKa = 10.85PLIHH70 pKa = 7.13EE71 pKa = 4.31PTDD74 pKa = 4.59PIWNDD79 pKa = 3.2VKK81 pKa = 11.05RR82 pKa = 11.84EE83 pKa = 3.86TLMKK87 pKa = 10.06IGSLFPQVQSLPFEE101 pKa = 4.61GGFDD105 pKa = 3.44HH106 pKa = 7.42VPFEE110 pKa = 4.53SSSSAGYY117 pKa = 10.46GYY119 pKa = 10.37DD120 pKa = 3.4GKK122 pKa = 10.94KK123 pKa = 10.68GEE125 pKa = 4.42GNNFHH130 pKa = 6.9RR131 pKa = 11.84AKK133 pKa = 10.48SIANAAVRR141 pKa = 11.84KK142 pKa = 9.46FSEE145 pKa = 5.34DD146 pKa = 2.92IDD148 pKa = 4.0NQGYY152 pKa = 10.12DD153 pKa = 3.49YY154 pKa = 11.34AVSHH158 pKa = 7.41LIQQGTPDD166 pKa = 3.07IAFTRR171 pKa = 11.84TQLAKK176 pKa = 10.59LPSIKK181 pKa = 10.16VRR183 pKa = 11.84IVFGEE188 pKa = 3.94AFHH191 pKa = 6.98NILIEE196 pKa = 4.29GLSAAPLLEE205 pKa = 4.0AFKK208 pKa = 11.16RR209 pKa = 11.84MDD211 pKa = 3.07TFYY214 pKa = 10.3FTGKK218 pKa = 10.29DD219 pKa = 3.08PTIYY223 pKa = 10.39VPRR226 pKa = 11.84ILHH229 pKa = 6.16KK230 pKa = 10.44MSTNEE235 pKa = 3.37GWFICLDD242 pKa = 3.51WKK244 pKa = 11.31AFDD247 pKa = 4.86ASVQLWEE254 pKa = 4.4IDD256 pKa = 3.43HH257 pKa = 6.94AFNCIQQLLAFPTEE271 pKa = 4.26LSRR274 pKa = 11.84LAFLFTRR281 pKa = 11.84EE282 pKa = 3.97SFKK285 pKa = 10.55QRR287 pKa = 11.84KK288 pKa = 8.16LADD291 pKa = 3.76PNGILWMRR299 pKa = 11.84KK300 pKa = 8.66GGIPSGSYY308 pKa = 6.73YY309 pKa = 10.66TNIIGSVINYY319 pKa = 8.88NRR321 pKa = 11.84IEE323 pKa = 4.26YY324 pKa = 8.74VCKK327 pKa = 10.45RR328 pKa = 11.84LGLQKK333 pKa = 9.64TSCYY337 pKa = 10.02VQGDD341 pKa = 3.77DD342 pKa = 4.79SLIHH346 pKa = 5.7ITGDD350 pKa = 3.28AKK352 pKa = 10.85PDD354 pKa = 3.48LTQLQMLGEE363 pKa = 4.15QFGWTLNIPKK373 pKa = 10.28CSLTQDD379 pKa = 3.44SQLVTFLGRR388 pKa = 11.84SQMHH392 pKa = 5.43QLNIRR397 pKa = 11.84EE398 pKa = 4.05RR399 pKa = 11.84LKK401 pKa = 10.86VLRR404 pKa = 11.84LMCFPEE410 pKa = 4.31YY411 pKa = 10.24KK412 pKa = 10.45VEE414 pKa = 4.15DD415 pKa = 4.08PKK417 pKa = 11.1ISTTRR422 pKa = 11.84VKK424 pKa = 10.77AIARR428 pKa = 11.84DD429 pKa = 3.94AGWSDD434 pKa = 3.14PVYY437 pKa = 11.24NKK439 pKa = 10.13IYY441 pKa = 10.3LQLKK445 pKa = 8.15RR446 pKa = 11.84LYY448 pKa = 10.91GEE450 pKa = 4.25VEE452 pKa = 4.08RR453 pKa = 11.84LPPHH457 pKa = 6.92LATFVDD463 pKa = 4.39RR464 pKa = 11.84FDD466 pKa = 4.17FQDD469 pKa = 3.14VNMM472 pKa = 4.97
MM1 pKa = 7.41EE2 pKa = 6.01AGLIEE7 pKa = 4.58IGNIPEE13 pKa = 3.69RR14 pKa = 11.84HH15 pKa = 6.05LRR17 pKa = 11.84DD18 pKa = 3.31EE19 pKa = 4.91FIILVDD25 pKa = 3.62QPAYY29 pKa = 10.87DD30 pKa = 3.58SVRR33 pKa = 11.84RR34 pKa = 11.84NAPQADD40 pKa = 3.77MQEE43 pKa = 3.77IDD45 pKa = 2.52GWARR49 pKa = 11.84SFYY52 pKa = 9.43TVEE55 pKa = 4.96GIMASIMQFSKK66 pKa = 10.85PLIHH70 pKa = 7.13EE71 pKa = 4.31PTDD74 pKa = 4.59PIWNDD79 pKa = 3.2VKK81 pKa = 11.05RR82 pKa = 11.84EE83 pKa = 3.86TLMKK87 pKa = 10.06IGSLFPQVQSLPFEE101 pKa = 4.61GGFDD105 pKa = 3.44HH106 pKa = 7.42VPFEE110 pKa = 4.53SSSSAGYY117 pKa = 10.46GYY119 pKa = 10.37DD120 pKa = 3.4GKK122 pKa = 10.94KK123 pKa = 10.68GEE125 pKa = 4.42GNNFHH130 pKa = 6.9RR131 pKa = 11.84AKK133 pKa = 10.48SIANAAVRR141 pKa = 11.84KK142 pKa = 9.46FSEE145 pKa = 5.34DD146 pKa = 2.92IDD148 pKa = 4.0NQGYY152 pKa = 10.12DD153 pKa = 3.49YY154 pKa = 11.34AVSHH158 pKa = 7.41LIQQGTPDD166 pKa = 3.07IAFTRR171 pKa = 11.84TQLAKK176 pKa = 10.59LPSIKK181 pKa = 10.16VRR183 pKa = 11.84IVFGEE188 pKa = 3.94AFHH191 pKa = 6.98NILIEE196 pKa = 4.29GLSAAPLLEE205 pKa = 4.0AFKK208 pKa = 11.16RR209 pKa = 11.84MDD211 pKa = 3.07TFYY214 pKa = 10.3FTGKK218 pKa = 10.29DD219 pKa = 3.08PTIYY223 pKa = 10.39VPRR226 pKa = 11.84ILHH229 pKa = 6.16KK230 pKa = 10.44MSTNEE235 pKa = 3.37GWFICLDD242 pKa = 3.51WKK244 pKa = 11.31AFDD247 pKa = 4.86ASVQLWEE254 pKa = 4.4IDD256 pKa = 3.43HH257 pKa = 6.94AFNCIQQLLAFPTEE271 pKa = 4.26LSRR274 pKa = 11.84LAFLFTRR281 pKa = 11.84EE282 pKa = 3.97SFKK285 pKa = 10.55QRR287 pKa = 11.84KK288 pKa = 8.16LADD291 pKa = 3.76PNGILWMRR299 pKa = 11.84KK300 pKa = 8.66GGIPSGSYY308 pKa = 6.73YY309 pKa = 10.66TNIIGSVINYY319 pKa = 8.88NRR321 pKa = 11.84IEE323 pKa = 4.26YY324 pKa = 8.74VCKK327 pKa = 10.45RR328 pKa = 11.84LGLQKK333 pKa = 9.64TSCYY337 pKa = 10.02VQGDD341 pKa = 3.77DD342 pKa = 4.79SLIHH346 pKa = 5.7ITGDD350 pKa = 3.28AKK352 pKa = 10.85PDD354 pKa = 3.48LTQLQMLGEE363 pKa = 4.15QFGWTLNIPKK373 pKa = 10.28CSLTQDD379 pKa = 3.44SQLVTFLGRR388 pKa = 11.84SQMHH392 pKa = 5.43QLNIRR397 pKa = 11.84EE398 pKa = 4.05RR399 pKa = 11.84LKK401 pKa = 10.86VLRR404 pKa = 11.84LMCFPEE410 pKa = 4.31YY411 pKa = 10.24KK412 pKa = 10.45VEE414 pKa = 4.15DD415 pKa = 4.08PKK417 pKa = 11.1ISTTRR422 pKa = 11.84VKK424 pKa = 10.77AIARR428 pKa = 11.84DD429 pKa = 3.94AGWSDD434 pKa = 3.14PVYY437 pKa = 11.24NKK439 pKa = 10.13IYY441 pKa = 10.3LQLKK445 pKa = 8.15RR446 pKa = 11.84LYY448 pKa = 10.91GEE450 pKa = 4.25VEE452 pKa = 4.08RR453 pKa = 11.84LPPHH457 pKa = 6.92LATFVDD463 pKa = 4.39RR464 pKa = 11.84FDD466 pKa = 4.17FQDD469 pKa = 3.14VNMM472 pKa = 4.97
Molecular weight: 53.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F4MST4|F4MST4_9VIRU Putative capsid protein OS=Fig cryptic virus OX=882768 GN=CP PE=4 SV=1
MM1 pKa = 7.75AAPNFDD7 pKa = 3.5ILEE10 pKa = 4.22DD11 pKa = 4.74LIADD15 pKa = 3.64GRR17 pKa = 11.84LQITPYY23 pKa = 9.12VTSTEE28 pKa = 3.95PKK30 pKa = 10.08IFDD33 pKa = 3.5VDD35 pKa = 6.02FDD37 pKa = 4.5ALWNRR42 pKa = 11.84INAIIVPIYY51 pKa = 9.43TNWDD55 pKa = 3.3QLRR58 pKa = 11.84FHH60 pKa = 7.04YY61 pKa = 10.3PGTTTTVTNNITLDD75 pKa = 3.42VGHH78 pKa = 7.46LVTFALWQLYY88 pKa = 9.63NRR90 pKa = 11.84LRR92 pKa = 11.84SVAEE96 pKa = 4.33CFHH99 pKa = 6.54HH100 pKa = 5.95PTRR103 pKa = 11.84TSRR106 pKa = 11.84MNTRR110 pKa = 11.84APISNRR116 pKa = 11.84MEE118 pKa = 4.22FPSFLSSMLEE128 pKa = 4.08SISWLRR134 pKa = 11.84INDD137 pKa = 3.73GPVDD141 pKa = 3.83YY142 pKa = 10.54LALFTAPTGTSNNYY156 pKa = 8.92GRR158 pKa = 11.84STVQVPDD165 pKa = 3.33NGCYY169 pKa = 10.21DD170 pKa = 3.83RR171 pKa = 11.84LTGKK175 pKa = 9.98LRR177 pKa = 11.84ALGVQLTPIDD187 pKa = 3.93MTPSAGSFWPTTQVITEE204 pKa = 4.11EE205 pKa = 3.92EE206 pKa = 4.84LYY208 pKa = 11.05NIVGTFHH215 pKa = 6.9SSHH218 pKa = 6.16YY219 pKa = 9.99VNEE222 pKa = 4.08DD223 pKa = 3.49AIRR226 pKa = 11.84AIFLAGTAHH235 pKa = 6.11SQPFPDD241 pKa = 4.19FGMTVGFVTDD251 pKa = 3.81RR252 pKa = 11.84TVMDD256 pKa = 4.83PLAALAAPTGTPRR269 pKa = 11.84SEE271 pKa = 4.13EE272 pKa = 3.87ATSAGRR278 pKa = 11.84PVHH281 pKa = 6.35GMYY284 pKa = 10.46NVNHH288 pKa = 7.02RR289 pKa = 11.84GIQPAAGEE297 pKa = 4.19QNPSGCYY304 pKa = 9.45IIGRR308 pKa = 11.84GYY310 pKa = 8.98EE311 pKa = 3.8RR312 pKa = 11.84RR313 pKa = 11.84YY314 pKa = 7.61TCYY317 pKa = 9.55LARR320 pKa = 11.84RR321 pKa = 11.84IAVFEE326 pKa = 4.2ANRR329 pKa = 11.84IIRR332 pKa = 11.84YY333 pKa = 9.49RR334 pKa = 11.84FMRR337 pKa = 4.14
MM1 pKa = 7.75AAPNFDD7 pKa = 3.5ILEE10 pKa = 4.22DD11 pKa = 4.74LIADD15 pKa = 3.64GRR17 pKa = 11.84LQITPYY23 pKa = 9.12VTSTEE28 pKa = 3.95PKK30 pKa = 10.08IFDD33 pKa = 3.5VDD35 pKa = 6.02FDD37 pKa = 4.5ALWNRR42 pKa = 11.84INAIIVPIYY51 pKa = 9.43TNWDD55 pKa = 3.3QLRR58 pKa = 11.84FHH60 pKa = 7.04YY61 pKa = 10.3PGTTTTVTNNITLDD75 pKa = 3.42VGHH78 pKa = 7.46LVTFALWQLYY88 pKa = 9.63NRR90 pKa = 11.84LRR92 pKa = 11.84SVAEE96 pKa = 4.33CFHH99 pKa = 6.54HH100 pKa = 5.95PTRR103 pKa = 11.84TSRR106 pKa = 11.84MNTRR110 pKa = 11.84APISNRR116 pKa = 11.84MEE118 pKa = 4.22FPSFLSSMLEE128 pKa = 4.08SISWLRR134 pKa = 11.84INDD137 pKa = 3.73GPVDD141 pKa = 3.83YY142 pKa = 10.54LALFTAPTGTSNNYY156 pKa = 8.92GRR158 pKa = 11.84STVQVPDD165 pKa = 3.33NGCYY169 pKa = 10.21DD170 pKa = 3.83RR171 pKa = 11.84LTGKK175 pKa = 9.98LRR177 pKa = 11.84ALGVQLTPIDD187 pKa = 3.93MTPSAGSFWPTTQVITEE204 pKa = 4.11EE205 pKa = 3.92EE206 pKa = 4.84LYY208 pKa = 11.05NIVGTFHH215 pKa = 6.9SSHH218 pKa = 6.16YY219 pKa = 9.99VNEE222 pKa = 4.08DD223 pKa = 3.49AIRR226 pKa = 11.84AIFLAGTAHH235 pKa = 6.11SQPFPDD241 pKa = 4.19FGMTVGFVTDD251 pKa = 3.81RR252 pKa = 11.84TVMDD256 pKa = 4.83PLAALAAPTGTPRR269 pKa = 11.84SEE271 pKa = 4.13EE272 pKa = 3.87ATSAGRR278 pKa = 11.84PVHH281 pKa = 6.35GMYY284 pKa = 10.46NVNHH288 pKa = 7.02RR289 pKa = 11.84GIQPAAGEE297 pKa = 4.19QNPSGCYY304 pKa = 9.45IIGRR308 pKa = 11.84GYY310 pKa = 8.98EE311 pKa = 3.8RR312 pKa = 11.84RR313 pKa = 11.84YY314 pKa = 7.61TCYY317 pKa = 9.55LARR320 pKa = 11.84RR321 pKa = 11.84IAVFEE326 pKa = 4.2ANRR329 pKa = 11.84IIRR332 pKa = 11.84YY333 pKa = 9.49RR334 pKa = 11.84FMRR337 pKa = 4.14
Molecular weight: 37.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
809 |
337 |
472 |
404.5 |
45.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.922 ± 0.619 | 1.236 ± 0.028 |
5.81 ± 0.434 | 4.944 ± 0.449 |
5.562 ± 0.294 | 6.551 ± 0.013 |
2.472 ± 0.113 | 7.417 ± 0.336 |
3.461 ± 1.628 | 8.282 ± 0.659 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.596 ± 0.042 | 4.574 ± 0.604 |
5.686 ± 0.478 | 4.203 ± 0.87 |
6.675 ± 0.759 | 5.933 ± 0.168 |
6.799 ± 1.7 | 5.315 ± 0.352 |
1.607 ± 0.07 | 3.956 ± 0.281 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
