
Beihai razor shell virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.8
Get precalculated fractions of proteins
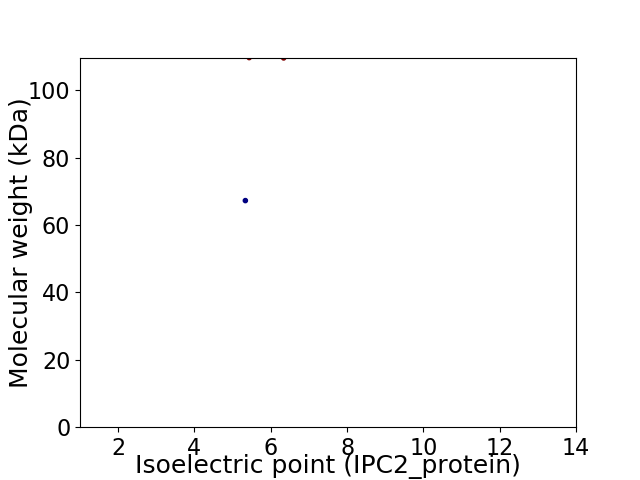
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
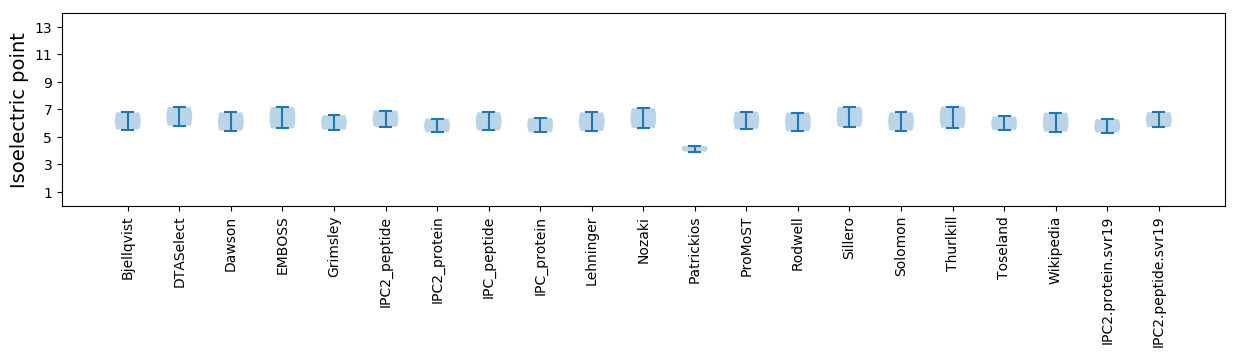
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KE88|A0A1L3KE88_9VIRU RNA-directed RNA polymerase OS=Beihai razor shell virus 3 OX=1922647 PE=4 SV=1
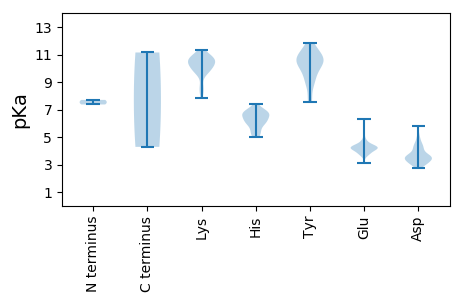
MM1 pKa = 7.7PEE3 pKa = 3.47GWQRR7 pKa = 11.84PPKK10 pKa = 10.35GKK12 pKa = 9.77YY13 pKa = 9.91ARR15 pKa = 11.84DD16 pKa = 3.38AMLGSLEE23 pKa = 3.84WHH25 pKa = 6.77LEE27 pKa = 3.71QNQRR31 pKa = 11.84ARR33 pKa = 11.84SQCQSVPTSALSDD46 pKa = 2.94IGGRR50 pKa = 11.84VAATYY55 pKa = 9.96CKK57 pKa = 9.71QQEE60 pKa = 4.4LCRR63 pKa = 11.84NLCQQGCWLQSASGEE78 pKa = 4.17PIAVRR83 pKa = 11.84KK84 pKa = 9.71GSRR87 pKa = 11.84GSQRR91 pKa = 11.84DD92 pKa = 3.76GAQHH96 pKa = 5.52QPVGQSFFSSEE107 pKa = 4.26AEE109 pKa = 4.15DD110 pKa = 4.34GCDD113 pKa = 3.21EE114 pKa = 5.15GVLQFLGFTGSGQRR128 pKa = 11.84IGPDD132 pKa = 3.0QRR134 pKa = 11.84FAEE137 pKa = 4.27INGAIACEE145 pKa = 4.24IEE147 pKa = 4.03HH148 pKa = 6.84HH149 pKa = 6.19EE150 pKa = 4.26CCKK153 pKa = 10.53TEE155 pKa = 3.92TEE157 pKa = 4.4EE158 pKa = 5.62RR159 pKa = 11.84EE160 pKa = 4.2CCQQVEE166 pKa = 4.31AGRR169 pKa = 11.84ALDD172 pKa = 3.69TEE174 pKa = 4.49RR175 pKa = 11.84VTRR178 pKa = 11.84LDD180 pKa = 3.99DD181 pKa = 3.5YY182 pKa = 11.85SLGFRR187 pKa = 11.84SLVSDD192 pKa = 4.2VARR195 pKa = 11.84SWGASGVDD203 pKa = 3.66DD204 pKa = 5.22AGVAATIDD212 pKa = 4.45EE213 pKa = 4.37IVATYY218 pKa = 9.57PCLRR222 pKa = 11.84DD223 pKa = 3.07AAVFGRR229 pKa = 11.84FLGSRR234 pKa = 11.84FGNEE238 pKa = 3.12RR239 pKa = 11.84VLNPLSTPGVEE250 pKa = 3.64WRR252 pKa = 11.84LFGADD257 pKa = 3.14NQTVFGYY264 pKa = 10.61DD265 pKa = 3.02AFFGEE270 pKa = 4.7YY271 pKa = 10.22NGVHH275 pKa = 6.1VEE277 pKa = 4.12DD278 pKa = 4.69LRR280 pKa = 11.84CAVIEE285 pKa = 4.5KK286 pKa = 10.05ILRR289 pKa = 11.84LIDD292 pKa = 4.18GPHH295 pKa = 7.37LDD297 pKa = 4.97RR298 pKa = 11.84IHH300 pKa = 6.76TFLKK304 pKa = 10.68DD305 pKa = 3.55EE306 pKa = 3.93PHH308 pKa = 6.49KK309 pKa = 10.25PEE311 pKa = 4.12KK312 pKa = 9.79VAEE315 pKa = 4.24GRR317 pKa = 11.84WRR319 pKa = 11.84IISGVSATDD328 pKa = 3.23QIACHH333 pKa = 6.33LLMGEE338 pKa = 4.3VLSSFSATPLTSGTAIGWNVMMHH361 pKa = 6.67GGLSSIFSLLPFGEE375 pKa = 4.25TGKK378 pKa = 10.71LVSADD383 pKa = 3.48RR384 pKa = 11.84KK385 pKa = 8.77SWDD388 pKa = 3.0WTVQKK393 pKa = 10.64LAVVLFTAFALLLLDD408 pKa = 5.8YY409 pKa = 8.66KK410 pKa = 8.18TQWEE414 pKa = 4.1RR415 pKa = 11.84KK416 pKa = 9.23AIVNHH421 pKa = 6.04ICSIFAIRR429 pKa = 11.84RR430 pKa = 11.84FDD432 pKa = 3.39CQGRR436 pKa = 11.84VVRR439 pKa = 11.84FDD441 pKa = 3.3NFGVLLSGWKK451 pKa = 8.28FTALFNSVVQHH462 pKa = 6.78LYY464 pKa = 11.29HH465 pKa = 6.39MLACSMLSLNGRR477 pKa = 11.84LPLSMGDD484 pKa = 3.39DD485 pKa = 3.77TVQEE489 pKa = 4.52EE490 pKa = 4.76MPQSYY495 pKa = 8.9WDD497 pKa = 3.51HH498 pKa = 6.62LATFGVILKK507 pKa = 8.62PTEE510 pKa = 4.01TFDD513 pKa = 4.67EE514 pKa = 4.83GFEE517 pKa = 4.11FCGAHH522 pKa = 5.92FKK524 pKa = 11.09VNSFSPVYY532 pKa = 10.13LPKK535 pKa = 10.16HH536 pKa = 6.09AYY538 pKa = 8.96NLRR541 pKa = 11.84DD542 pKa = 3.58VSPEE546 pKa = 3.81NLPALLISYY555 pKa = 9.39RR556 pKa = 11.84YY557 pKa = 10.42LYY559 pKa = 10.34MFDD562 pKa = 3.18EE563 pKa = 4.67RR564 pKa = 11.84RR565 pKa = 11.84FNALNRR571 pKa = 11.84WACEE575 pKa = 3.79MGFDD579 pKa = 4.27SLDD582 pKa = 3.43RR583 pKa = 11.84SEE585 pKa = 6.3LIFKK589 pKa = 10.28VLGVSEE595 pKa = 4.32MSEE598 pKa = 4.17VMVV601 pKa = 4.28
MM1 pKa = 7.7PEE3 pKa = 3.47GWQRR7 pKa = 11.84PPKK10 pKa = 10.35GKK12 pKa = 9.77YY13 pKa = 9.91ARR15 pKa = 11.84DD16 pKa = 3.38AMLGSLEE23 pKa = 3.84WHH25 pKa = 6.77LEE27 pKa = 3.71QNQRR31 pKa = 11.84ARR33 pKa = 11.84SQCQSVPTSALSDD46 pKa = 2.94IGGRR50 pKa = 11.84VAATYY55 pKa = 9.96CKK57 pKa = 9.71QQEE60 pKa = 4.4LCRR63 pKa = 11.84NLCQQGCWLQSASGEE78 pKa = 4.17PIAVRR83 pKa = 11.84KK84 pKa = 9.71GSRR87 pKa = 11.84GSQRR91 pKa = 11.84DD92 pKa = 3.76GAQHH96 pKa = 5.52QPVGQSFFSSEE107 pKa = 4.26AEE109 pKa = 4.15DD110 pKa = 4.34GCDD113 pKa = 3.21EE114 pKa = 5.15GVLQFLGFTGSGQRR128 pKa = 11.84IGPDD132 pKa = 3.0QRR134 pKa = 11.84FAEE137 pKa = 4.27INGAIACEE145 pKa = 4.24IEE147 pKa = 4.03HH148 pKa = 6.84HH149 pKa = 6.19EE150 pKa = 4.26CCKK153 pKa = 10.53TEE155 pKa = 3.92TEE157 pKa = 4.4EE158 pKa = 5.62RR159 pKa = 11.84EE160 pKa = 4.2CCQQVEE166 pKa = 4.31AGRR169 pKa = 11.84ALDD172 pKa = 3.69TEE174 pKa = 4.49RR175 pKa = 11.84VTRR178 pKa = 11.84LDD180 pKa = 3.99DD181 pKa = 3.5YY182 pKa = 11.85SLGFRR187 pKa = 11.84SLVSDD192 pKa = 4.2VARR195 pKa = 11.84SWGASGVDD203 pKa = 3.66DD204 pKa = 5.22AGVAATIDD212 pKa = 4.45EE213 pKa = 4.37IVATYY218 pKa = 9.57PCLRR222 pKa = 11.84DD223 pKa = 3.07AAVFGRR229 pKa = 11.84FLGSRR234 pKa = 11.84FGNEE238 pKa = 3.12RR239 pKa = 11.84VLNPLSTPGVEE250 pKa = 3.64WRR252 pKa = 11.84LFGADD257 pKa = 3.14NQTVFGYY264 pKa = 10.61DD265 pKa = 3.02AFFGEE270 pKa = 4.7YY271 pKa = 10.22NGVHH275 pKa = 6.1VEE277 pKa = 4.12DD278 pKa = 4.69LRR280 pKa = 11.84CAVIEE285 pKa = 4.5KK286 pKa = 10.05ILRR289 pKa = 11.84LIDD292 pKa = 4.18GPHH295 pKa = 7.37LDD297 pKa = 4.97RR298 pKa = 11.84IHH300 pKa = 6.76TFLKK304 pKa = 10.68DD305 pKa = 3.55EE306 pKa = 3.93PHH308 pKa = 6.49KK309 pKa = 10.25PEE311 pKa = 4.12KK312 pKa = 9.79VAEE315 pKa = 4.24GRR317 pKa = 11.84WRR319 pKa = 11.84IISGVSATDD328 pKa = 3.23QIACHH333 pKa = 6.33LLMGEE338 pKa = 4.3VLSSFSATPLTSGTAIGWNVMMHH361 pKa = 6.67GGLSSIFSLLPFGEE375 pKa = 4.25TGKK378 pKa = 10.71LVSADD383 pKa = 3.48RR384 pKa = 11.84KK385 pKa = 8.77SWDD388 pKa = 3.0WTVQKK393 pKa = 10.64LAVVLFTAFALLLLDD408 pKa = 5.8YY409 pKa = 8.66KK410 pKa = 8.18TQWEE414 pKa = 4.1RR415 pKa = 11.84KK416 pKa = 9.23AIVNHH421 pKa = 6.04ICSIFAIRR429 pKa = 11.84RR430 pKa = 11.84FDD432 pKa = 3.39CQGRR436 pKa = 11.84VVRR439 pKa = 11.84FDD441 pKa = 3.3NFGVLLSGWKK451 pKa = 8.28FTALFNSVVQHH462 pKa = 6.78LYY464 pKa = 11.29HH465 pKa = 6.39MLACSMLSLNGRR477 pKa = 11.84LPLSMGDD484 pKa = 3.39DD485 pKa = 3.77TVQEE489 pKa = 4.52EE490 pKa = 4.76MPQSYY495 pKa = 8.9WDD497 pKa = 3.51HH498 pKa = 6.62LATFGVILKK507 pKa = 8.62PTEE510 pKa = 4.01TFDD513 pKa = 4.67EE514 pKa = 4.83GFEE517 pKa = 4.11FCGAHH522 pKa = 5.92FKK524 pKa = 11.09VNSFSPVYY532 pKa = 10.13LPKK535 pKa = 10.16HH536 pKa = 6.09AYY538 pKa = 8.96NLRR541 pKa = 11.84DD542 pKa = 3.58VSPEE546 pKa = 3.81NLPALLISYY555 pKa = 9.39RR556 pKa = 11.84YY557 pKa = 10.42LYY559 pKa = 10.34MFDD562 pKa = 3.18EE563 pKa = 4.67RR564 pKa = 11.84RR565 pKa = 11.84FNALNRR571 pKa = 11.84WACEE575 pKa = 3.79MGFDD579 pKa = 4.27SLDD582 pKa = 3.43RR583 pKa = 11.84SEE585 pKa = 6.3LIFKK589 pKa = 10.28VLGVSEE595 pKa = 4.32MSEE598 pKa = 4.17VMVV601 pKa = 4.28
Molecular weight: 67.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE88|A0A1L3KE88_9VIRU RNA-directed RNA polymerase OS=Beihai razor shell virus 3 OX=1922647 PE=4 SV=1
MM1 pKa = 7.37KK2 pKa = 10.55RR3 pKa = 11.84LGLLLTMTFVEE14 pKa = 4.41QTEE17 pKa = 4.55HH18 pKa = 6.97VFEE21 pKa = 4.23AVRR24 pKa = 11.84NKK26 pKa = 9.93SRR28 pKa = 11.84ALINTRR34 pKa = 11.84LSEE37 pKa = 4.06IWNMTFPEE45 pKa = 3.82IRR47 pKa = 11.84LVEE50 pKa = 4.25RR51 pKa = 11.84KK52 pKa = 8.52PWEE55 pKa = 4.49SVSSTGDD62 pKa = 3.16EE63 pKa = 4.33SVLHH67 pKa = 5.2QFCMSACFVIIALFVLPVICCIVRR91 pKa = 11.84SVGAVAWDD99 pKa = 3.87HH100 pKa = 6.37LCSFALVMVEE110 pKa = 3.77LVKK113 pKa = 10.66WMLRR117 pKa = 11.84VTVAVTTVIMLVVRR131 pKa = 11.84LVWAWWYY138 pKa = 11.11NDD140 pKa = 4.78LYY142 pKa = 11.41SCMIATFLVAVVSIYY157 pKa = 10.81LLWRR161 pKa = 11.84LFEE164 pKa = 5.33ADD166 pKa = 2.92SAQIYY171 pKa = 10.69SGFEE175 pKa = 4.06SIKK178 pKa = 10.78GGTNAPFRR186 pKa = 11.84IEE188 pKa = 4.26EE189 pKa = 4.17LSIMPLGSSDD199 pKa = 4.42FEE201 pKa = 4.39ASSPSSSVVQVGFQRR216 pKa = 11.84GFGLNRR222 pKa = 11.84AFVTVGNAIYY232 pKa = 10.12FRR234 pKa = 11.84EE235 pKa = 4.34LDD237 pKa = 3.61GKK239 pKa = 10.2HH240 pKa = 5.47YY241 pKa = 10.59FSVPTHH247 pKa = 6.63VITALSSVTSAVPLLKK263 pKa = 10.55RR264 pKa = 11.84GTTVIAVEE272 pKa = 4.68RR273 pKa = 11.84ICTLGLQHH281 pKa = 6.91LAEE284 pKa = 4.27FTVLVAGQRR293 pKa = 11.84ARR295 pKa = 11.84GMASLARR302 pKa = 11.84VGHH305 pKa = 5.02VDD307 pKa = 4.33RR308 pKa = 11.84RR309 pKa = 11.84DD310 pKa = 3.26ACPVVVHH317 pKa = 6.44AHH319 pKa = 5.9TDD321 pKa = 3.23SGSALSKK328 pKa = 10.78GQLLPATGMGSSGVHH343 pKa = 5.1VFHH346 pKa = 7.04HH347 pKa = 6.04TASTDD352 pKa = 3.51KK353 pKa = 10.11GWSGGLVTMDD363 pKa = 3.56GKK365 pKa = 10.92AIGQHH370 pKa = 5.37VGWVSAKK377 pKa = 10.49DD378 pKa = 3.77PYY380 pKa = 10.74HH381 pKa = 6.81GEE383 pKa = 4.0SKK385 pKa = 11.34ANVAFNLAYY394 pKa = 10.56LYY396 pKa = 10.78AIVDD400 pKa = 3.56WKK402 pKa = 10.78LRR404 pKa = 11.84PAGDD408 pKa = 3.56RR409 pKa = 11.84SGLEE413 pKa = 3.84ADD415 pKa = 4.54SFTSPLIDD423 pKa = 3.7RR424 pKa = 11.84LVADD428 pKa = 4.44YY429 pKa = 7.83EE430 pKa = 4.32TDD432 pKa = 5.03DD433 pKa = 4.25RR434 pKa = 11.84NDD436 pKa = 3.05WYY438 pKa = 11.35VEE440 pKa = 3.91EE441 pKa = 4.81LVSPNLDD448 pKa = 2.77VTYY451 pKa = 10.51IVFNRR456 pKa = 11.84RR457 pKa = 11.84SGKK460 pKa = 7.82VSRR463 pKa = 11.84YY464 pKa = 9.38EE465 pKa = 4.08RR466 pKa = 11.84DD467 pKa = 3.53DD468 pKa = 4.04LSSDD472 pKa = 3.0KK473 pKa = 11.31SSMIRR478 pKa = 11.84RR479 pKa = 11.84YY480 pKa = 9.34IAGSGTEE487 pKa = 3.76NAEE490 pKa = 4.05FQDD493 pKa = 4.08ASSEE497 pKa = 4.06MTTACEE503 pKa = 3.88EE504 pKa = 4.42YY505 pKa = 10.3VDD507 pKa = 3.52KK508 pKa = 10.72EE509 pKa = 3.68YY510 pKa = 11.12RR511 pKa = 11.84RR512 pKa = 11.84LVRR515 pKa = 11.84EE516 pKa = 3.6IDD518 pKa = 3.64EE519 pKa = 4.78LMGSYY524 pKa = 8.79MRR526 pKa = 11.84NNGVDD531 pKa = 2.91AAMDD535 pKa = 3.28VMRR538 pKa = 11.84EE539 pKa = 3.59ITYY542 pKa = 9.81LAKK545 pKa = 10.61AQSTYY550 pKa = 10.75IQTLCDD556 pKa = 3.52NLRR559 pKa = 11.84TPNRR563 pKa = 11.84GMEE566 pKa = 4.44ALPVVPVDD574 pKa = 3.49YY575 pKa = 11.45GNLRR579 pKa = 11.84TMMVLVEE586 pKa = 4.35QYY588 pKa = 11.79ADD590 pKa = 3.13IQRR593 pKa = 11.84QLGSMAATEE602 pKa = 3.82QLAAGEE608 pKa = 3.85IRR610 pKa = 11.84ARR612 pKa = 11.84VAAGEE617 pKa = 4.22PKK619 pKa = 10.31EE620 pKa = 4.19QVARR624 pKa = 11.84DD625 pKa = 3.56QMKK628 pKa = 10.2IIANMRR634 pKa = 11.84AINAAIEE641 pKa = 4.06LCPNIGRR648 pKa = 11.84EE649 pKa = 4.14LTMLARR655 pKa = 11.84NQAQLSADD663 pKa = 3.32EE664 pKa = 4.37VTSRR668 pKa = 11.84NSMDD672 pKa = 3.12RR673 pKa = 11.84VRR675 pKa = 11.84RR676 pKa = 11.84KK677 pKa = 9.77LRR679 pKa = 11.84NEE681 pKa = 4.6GYY683 pKa = 10.73DD684 pKa = 3.54PAADD688 pKa = 3.93PATIDD693 pKa = 3.4AVIAQRR699 pKa = 11.84QALLNAQQAAKK710 pKa = 10.22AIPLRR715 pKa = 11.84PEE717 pKa = 4.05APLVIPVAAQGPEE730 pKa = 4.25SLTLDD735 pKa = 3.48YY736 pKa = 10.99YY737 pKa = 10.78RR738 pKa = 11.84RR739 pKa = 11.84KK740 pKa = 9.68SQEE743 pKa = 3.7GTNLTGFNVYY753 pKa = 10.25FDD755 pKa = 4.02PLKK758 pKa = 10.61VDD760 pKa = 3.34EE761 pKa = 5.05EE762 pKa = 4.3EE763 pKa = 4.46VIRR766 pKa = 11.84QIRR769 pKa = 11.84DD770 pKa = 3.0IKK772 pKa = 10.47QSAVSFLVPRR782 pKa = 11.84PASSGLNSEE791 pKa = 4.17VLKK794 pKa = 10.87PKK796 pKa = 10.06VAQTMPASASLSSVTQSTSAEE817 pKa = 4.17EE818 pKa = 4.38TTKK821 pKa = 11.01SPTTLPNYY829 pKa = 7.55SQCQKK834 pKa = 10.84AGSDD838 pKa = 3.54PRR840 pKa = 11.84KK841 pKa = 10.12ASMPEE846 pKa = 4.22TQCLGALNGTLSRR859 pKa = 11.84TNGLEE864 pKa = 4.0ANVNQCLPVHH874 pKa = 6.01YY875 pKa = 7.56PTLEE879 pKa = 4.3AEE881 pKa = 4.44SQQLIANSKK890 pKa = 10.26SCVEE894 pKa = 4.19TCASKK899 pKa = 10.92GVGCSQPQASQSPLEE914 pKa = 4.14KK915 pKa = 10.71VRR917 pKa = 11.84VDD919 pKa = 3.7LSEE922 pKa = 3.72MARR925 pKa = 11.84NINRR929 pKa = 11.84LANPSSAPKK938 pKa = 10.52LKK940 pKa = 9.68MGVMRR945 pKa = 11.84EE946 pKa = 4.18FFSSLASQEE955 pKa = 3.94VDD957 pKa = 4.25SEE959 pKa = 4.2LDD961 pKa = 3.47QIKK964 pKa = 10.4GLQKK968 pKa = 11.05SMEE971 pKa = 4.04QLHH974 pKa = 6.73AKK976 pKa = 9.68LNTMSAAKK984 pKa = 9.73PKK986 pKa = 10.11RR987 pKa = 11.84RR988 pKa = 11.84NASVVSKK995 pKa = 11.15
MM1 pKa = 7.37KK2 pKa = 10.55RR3 pKa = 11.84LGLLLTMTFVEE14 pKa = 4.41QTEE17 pKa = 4.55HH18 pKa = 6.97VFEE21 pKa = 4.23AVRR24 pKa = 11.84NKK26 pKa = 9.93SRR28 pKa = 11.84ALINTRR34 pKa = 11.84LSEE37 pKa = 4.06IWNMTFPEE45 pKa = 3.82IRR47 pKa = 11.84LVEE50 pKa = 4.25RR51 pKa = 11.84KK52 pKa = 8.52PWEE55 pKa = 4.49SVSSTGDD62 pKa = 3.16EE63 pKa = 4.33SVLHH67 pKa = 5.2QFCMSACFVIIALFVLPVICCIVRR91 pKa = 11.84SVGAVAWDD99 pKa = 3.87HH100 pKa = 6.37LCSFALVMVEE110 pKa = 3.77LVKK113 pKa = 10.66WMLRR117 pKa = 11.84VTVAVTTVIMLVVRR131 pKa = 11.84LVWAWWYY138 pKa = 11.11NDD140 pKa = 4.78LYY142 pKa = 11.41SCMIATFLVAVVSIYY157 pKa = 10.81LLWRR161 pKa = 11.84LFEE164 pKa = 5.33ADD166 pKa = 2.92SAQIYY171 pKa = 10.69SGFEE175 pKa = 4.06SIKK178 pKa = 10.78GGTNAPFRR186 pKa = 11.84IEE188 pKa = 4.26EE189 pKa = 4.17LSIMPLGSSDD199 pKa = 4.42FEE201 pKa = 4.39ASSPSSSVVQVGFQRR216 pKa = 11.84GFGLNRR222 pKa = 11.84AFVTVGNAIYY232 pKa = 10.12FRR234 pKa = 11.84EE235 pKa = 4.34LDD237 pKa = 3.61GKK239 pKa = 10.2HH240 pKa = 5.47YY241 pKa = 10.59FSVPTHH247 pKa = 6.63VITALSSVTSAVPLLKK263 pKa = 10.55RR264 pKa = 11.84GTTVIAVEE272 pKa = 4.68RR273 pKa = 11.84ICTLGLQHH281 pKa = 6.91LAEE284 pKa = 4.27FTVLVAGQRR293 pKa = 11.84ARR295 pKa = 11.84GMASLARR302 pKa = 11.84VGHH305 pKa = 5.02VDD307 pKa = 4.33RR308 pKa = 11.84RR309 pKa = 11.84DD310 pKa = 3.26ACPVVVHH317 pKa = 6.44AHH319 pKa = 5.9TDD321 pKa = 3.23SGSALSKK328 pKa = 10.78GQLLPATGMGSSGVHH343 pKa = 5.1VFHH346 pKa = 7.04HH347 pKa = 6.04TASTDD352 pKa = 3.51KK353 pKa = 10.11GWSGGLVTMDD363 pKa = 3.56GKK365 pKa = 10.92AIGQHH370 pKa = 5.37VGWVSAKK377 pKa = 10.49DD378 pKa = 3.77PYY380 pKa = 10.74HH381 pKa = 6.81GEE383 pKa = 4.0SKK385 pKa = 11.34ANVAFNLAYY394 pKa = 10.56LYY396 pKa = 10.78AIVDD400 pKa = 3.56WKK402 pKa = 10.78LRR404 pKa = 11.84PAGDD408 pKa = 3.56RR409 pKa = 11.84SGLEE413 pKa = 3.84ADD415 pKa = 4.54SFTSPLIDD423 pKa = 3.7RR424 pKa = 11.84LVADD428 pKa = 4.44YY429 pKa = 7.83EE430 pKa = 4.32TDD432 pKa = 5.03DD433 pKa = 4.25RR434 pKa = 11.84NDD436 pKa = 3.05WYY438 pKa = 11.35VEE440 pKa = 3.91EE441 pKa = 4.81LVSPNLDD448 pKa = 2.77VTYY451 pKa = 10.51IVFNRR456 pKa = 11.84RR457 pKa = 11.84SGKK460 pKa = 7.82VSRR463 pKa = 11.84YY464 pKa = 9.38EE465 pKa = 4.08RR466 pKa = 11.84DD467 pKa = 3.53DD468 pKa = 4.04LSSDD472 pKa = 3.0KK473 pKa = 11.31SSMIRR478 pKa = 11.84RR479 pKa = 11.84YY480 pKa = 9.34IAGSGTEE487 pKa = 3.76NAEE490 pKa = 4.05FQDD493 pKa = 4.08ASSEE497 pKa = 4.06MTTACEE503 pKa = 3.88EE504 pKa = 4.42YY505 pKa = 10.3VDD507 pKa = 3.52KK508 pKa = 10.72EE509 pKa = 3.68YY510 pKa = 11.12RR511 pKa = 11.84RR512 pKa = 11.84LVRR515 pKa = 11.84EE516 pKa = 3.6IDD518 pKa = 3.64EE519 pKa = 4.78LMGSYY524 pKa = 8.79MRR526 pKa = 11.84NNGVDD531 pKa = 2.91AAMDD535 pKa = 3.28VMRR538 pKa = 11.84EE539 pKa = 3.59ITYY542 pKa = 9.81LAKK545 pKa = 10.61AQSTYY550 pKa = 10.75IQTLCDD556 pKa = 3.52NLRR559 pKa = 11.84TPNRR563 pKa = 11.84GMEE566 pKa = 4.44ALPVVPVDD574 pKa = 3.49YY575 pKa = 11.45GNLRR579 pKa = 11.84TMMVLVEE586 pKa = 4.35QYY588 pKa = 11.79ADD590 pKa = 3.13IQRR593 pKa = 11.84QLGSMAATEE602 pKa = 3.82QLAAGEE608 pKa = 3.85IRR610 pKa = 11.84ARR612 pKa = 11.84VAAGEE617 pKa = 4.22PKK619 pKa = 10.31EE620 pKa = 4.19QVARR624 pKa = 11.84DD625 pKa = 3.56QMKK628 pKa = 10.2IIANMRR634 pKa = 11.84AINAAIEE641 pKa = 4.06LCPNIGRR648 pKa = 11.84EE649 pKa = 4.14LTMLARR655 pKa = 11.84NQAQLSADD663 pKa = 3.32EE664 pKa = 4.37VTSRR668 pKa = 11.84NSMDD672 pKa = 3.12RR673 pKa = 11.84VRR675 pKa = 11.84RR676 pKa = 11.84KK677 pKa = 9.77LRR679 pKa = 11.84NEE681 pKa = 4.6GYY683 pKa = 10.73DD684 pKa = 3.54PAADD688 pKa = 3.93PATIDD693 pKa = 3.4AVIAQRR699 pKa = 11.84QALLNAQQAAKK710 pKa = 10.22AIPLRR715 pKa = 11.84PEE717 pKa = 4.05APLVIPVAAQGPEE730 pKa = 4.25SLTLDD735 pKa = 3.48YY736 pKa = 10.99YY737 pKa = 10.78RR738 pKa = 11.84RR739 pKa = 11.84KK740 pKa = 9.68SQEE743 pKa = 3.7GTNLTGFNVYY753 pKa = 10.25FDD755 pKa = 4.02PLKK758 pKa = 10.61VDD760 pKa = 3.34EE761 pKa = 5.05EE762 pKa = 4.3EE763 pKa = 4.46VIRR766 pKa = 11.84QIRR769 pKa = 11.84DD770 pKa = 3.0IKK772 pKa = 10.47QSAVSFLVPRR782 pKa = 11.84PASSGLNSEE791 pKa = 4.17VLKK794 pKa = 10.87PKK796 pKa = 10.06VAQTMPASASLSSVTQSTSAEE817 pKa = 4.17EE818 pKa = 4.38TTKK821 pKa = 11.01SPTTLPNYY829 pKa = 7.55SQCQKK834 pKa = 10.84AGSDD838 pKa = 3.54PRR840 pKa = 11.84KK841 pKa = 10.12ASMPEE846 pKa = 4.22TQCLGALNGTLSRR859 pKa = 11.84TNGLEE864 pKa = 4.0ANVNQCLPVHH874 pKa = 6.01YY875 pKa = 7.56PTLEE879 pKa = 4.3AEE881 pKa = 4.44SQQLIANSKK890 pKa = 10.26SCVEE894 pKa = 4.19TCASKK899 pKa = 10.92GVGCSQPQASQSPLEE914 pKa = 4.14KK915 pKa = 10.71VRR917 pKa = 11.84VDD919 pKa = 3.7LSEE922 pKa = 3.72MARR925 pKa = 11.84NINRR929 pKa = 11.84LANPSSAPKK938 pKa = 10.52LKK940 pKa = 9.68MGVMRR945 pKa = 11.84EE946 pKa = 4.18FFSSLASQEE955 pKa = 3.94VDD957 pKa = 4.25SEE959 pKa = 4.2LDD961 pKa = 3.47QIKK964 pKa = 10.4GLQKK968 pKa = 11.05SMEE971 pKa = 4.04QLHH974 pKa = 6.73AKK976 pKa = 9.68LNTMSAAKK984 pKa = 9.73PKK986 pKa = 10.11RR987 pKa = 11.84RR988 pKa = 11.84NASVVSKK995 pKa = 11.15
Molecular weight: 109.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1596 |
601 |
995 |
798.0 |
88.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.023 ± 0.801 | 2.256 ± 0.473 |
5.013 ± 0.336 | 6.454 ± 0.192 |
3.947 ± 0.979 | 6.642 ± 0.962 |
2.005 ± 0.343 | 4.261 ± 0.139 |
3.697 ± 0.279 | 9.273 ± 0.197 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.882 ± 0.375 | 3.509 ± 0.355 |
4.01 ± 0.182 | 4.386 ± 0.056 |
6.642 ± 0.007 | 8.584 ± 0.485 |
5.013 ± 0.445 | 8.271 ± 0.669 |
1.566 ± 0.311 | 2.569 ± 0.125 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
