
Rhodobacter maris
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Rhodobacter
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
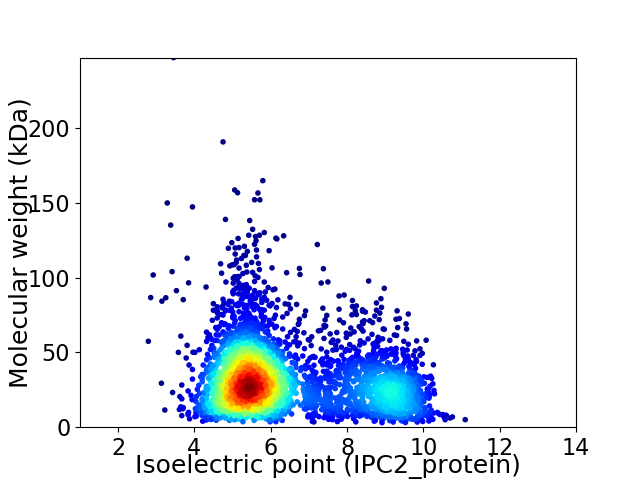
Virtual 2D-PAGE plot for 3687 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A285SG54|A0A285SG54_9RHOB Myo-inositol-1(Or 4)-monophosphatase OS=Rhodobacter maris OX=446682 GN=SAMN05877831_10551 PE=4 SV=1
MM1 pKa = 6.33TTWTIQGFSGSDD13 pKa = 3.29LIIAEE18 pKa = 4.58GGSTLSVGSNFMLDD32 pKa = 3.84PTWDD36 pKa = 3.35ASTDD40 pKa = 3.24ATSYY44 pKa = 10.63TFTDD48 pKa = 3.79PDD50 pKa = 3.75DD51 pKa = 4.25TNLSGDD57 pKa = 3.75NLNNEE62 pKa = 4.64HH63 pKa = 6.9GNDD66 pKa = 3.68GNQTINVYY74 pKa = 10.42DD75 pKa = 3.48SSGTLIHH82 pKa = 6.61SGQVYY87 pKa = 10.17IEE89 pKa = 4.64DD90 pKa = 3.75SAVFTAPDD98 pKa = 3.86GSSITLYY105 pKa = 9.16TLEE108 pKa = 4.14VNGVVVGTVTSAPLVPGVTYY128 pKa = 10.47HH129 pKa = 5.02VTAINDD135 pKa = 3.48VTTGPLYY142 pKa = 10.16TALASTTYY150 pKa = 11.08DD151 pKa = 3.42PDD153 pKa = 3.47LANTVTGGDD162 pKa = 3.71YY163 pKa = 11.07ADD165 pKa = 5.22SITGGAGNDD174 pKa = 4.08TITTGAGSDD183 pKa = 3.88TIDD186 pKa = 3.66GGTGNDD192 pKa = 3.7SIQFGTGGDD201 pKa = 3.87SVHH204 pKa = 6.93GGDD207 pKa = 4.03GNDD210 pKa = 4.42YY211 pKa = 10.44IDD213 pKa = 5.72DD214 pKa = 4.15YY215 pKa = 11.62AGSTGYY221 pKa = 10.74VYY223 pKa = 11.24NDD225 pKa = 3.72TLYY228 pKa = 11.47GDD230 pKa = 4.74AGNDD234 pKa = 3.62TIFAGSGNDD243 pKa = 3.77YY244 pKa = 11.27VDD246 pKa = 4.45GGADD250 pKa = 3.21NDD252 pKa = 4.06NIYY255 pKa = 11.21GEE257 pKa = 5.03DD258 pKa = 4.07GNDD261 pKa = 3.51TLLGGAGCDD270 pKa = 3.6WIEE273 pKa = 4.35GGAGADD279 pKa = 4.12SIDD282 pKa = 3.92GGTGTDD288 pKa = 3.86IILGGDD294 pKa = 3.75GNDD297 pKa = 4.2TIHH300 pKa = 6.85GGDD303 pKa = 3.56GADD306 pKa = 3.37YY307 pKa = 10.94LAGNTGNDD315 pKa = 3.35VLYY318 pKa = 11.21GEE320 pKa = 5.24ADD322 pKa = 3.17ADD324 pKa = 3.54RR325 pKa = 11.84FYY327 pKa = 11.43FEE329 pKa = 5.42NGWGSDD335 pKa = 3.21TVYY338 pKa = 11.0GGSTTTAGGVDD349 pKa = 5.12LDD351 pKa = 4.4TLDD354 pKa = 4.06FSYY357 pKa = 9.84DD358 pKa = 3.48TVSGVSVTFTDD369 pKa = 3.8WEE371 pKa = 4.95DD372 pKa = 3.38GTATDD377 pKa = 4.0GTSTVTFDD385 pKa = 4.25NIEE388 pKa = 4.51GIYY391 pKa = 9.1GTAFGDD397 pKa = 4.06TINASADD404 pKa = 3.51GSGLTLDD411 pKa = 4.2GQAGNDD417 pKa = 3.94SIIGGSGDD425 pKa = 3.75DD426 pKa = 4.74CILGGAGDD434 pKa = 4.0DD435 pKa = 4.23TIFSGFGNDD444 pKa = 3.01TVYY447 pKa = 11.14GGDD450 pKa = 3.89GNDD453 pKa = 4.59SIDD456 pKa = 4.07DD457 pKa = 3.75KK458 pKa = 11.64SGVLAEE464 pKa = 4.52TYY466 pKa = 10.07TNYY469 pKa = 10.09VDD471 pKa = 5.24AGAGNDD477 pKa = 3.64TVFTGGGSDD486 pKa = 3.71TILGGEE492 pKa = 4.92GDD494 pKa = 4.34DD495 pKa = 4.3QLHH498 pKa = 6.64AEE500 pKa = 4.52AGDD503 pKa = 3.63DD504 pKa = 3.99LVYY507 pKa = 11.03GGAGADD513 pKa = 3.63TLWGNEE519 pKa = 4.11GADD522 pKa = 3.97TLWGGDD528 pKa = 3.5GADD531 pKa = 4.53SILGGIGDD539 pKa = 4.05DD540 pKa = 5.08LIYY543 pKa = 11.25GEE545 pKa = 5.27AGNDD549 pKa = 3.82TITGGDD555 pKa = 3.62GADD558 pKa = 4.16SISGGADD565 pKa = 2.9NDD567 pKa = 4.27VITGDD572 pKa = 4.74LGDD575 pKa = 4.26DD576 pKa = 3.71TLAGGTGADD585 pKa = 3.73TLTGGTGADD594 pKa = 3.59TYY596 pKa = 11.79VMSDD600 pKa = 2.87GDD602 pKa = 4.01GADD605 pKa = 3.24TVTDD609 pKa = 3.91FDD611 pKa = 3.72MTMVSGLTTDD621 pKa = 3.29QFDD624 pKa = 3.74VSGLKK629 pKa = 10.18DD630 pKa = 3.42AEE632 pKa = 4.28GNPVNVWDD640 pKa = 3.92VTVDD644 pKa = 3.33SDD646 pKa = 3.85EE647 pKa = 4.14QGNAVLFFPDD657 pKa = 3.75GTSVTLIGVSKK668 pKa = 9.65TSVNTAPMLHH678 pKa = 6.68AMGIPCLVAGSPVATPRR695 pKa = 11.84GPVAVEE701 pKa = 3.42QLVPGDD707 pKa = 3.93LVITASGAAEE717 pKa = 3.74PVLWTGARR725 pKa = 11.84AVTAAEE731 pKa = 4.03MAADD735 pKa = 3.88PRR737 pKa = 11.84LLPVEE742 pKa = 4.51IGAGRR747 pKa = 11.84LGNAAPVRR755 pKa = 11.84LSALHH760 pKa = 6.12AVHH763 pKa = 6.47VPARR767 pKa = 11.84GGALARR773 pKa = 11.84AGHH776 pKa = 5.87MAATGWGGARR786 pKa = 11.84ILHH789 pKa = 5.67GLARR793 pKa = 11.84QEE795 pKa = 3.63AGLRR799 pKa = 11.84YY800 pKa = 9.25HH801 pKa = 6.89HH802 pKa = 7.04LLLPRR807 pKa = 11.84HH808 pKa = 5.83ALISVAGLWVEE819 pKa = 4.26SFWPGRR825 pKa = 11.84QGLAGLDD832 pKa = 3.5PASRR836 pKa = 11.84TRR838 pKa = 11.84LIRR841 pKa = 11.84ARR843 pKa = 11.84PQLAQVLWGGASVAACYY860 pKa = 9.04SQPAAPFLRR869 pKa = 11.84RR870 pKa = 11.84RR871 pKa = 11.84EE872 pKa = 3.9IDD874 pKa = 3.38RR875 pKa = 11.84AACSNWSLLARR886 pKa = 11.84EE887 pKa = 4.9KK888 pKa = 10.36PHH890 pKa = 7.43SDD892 pKa = 2.79GFQASSGQASTLL904 pKa = 3.57
MM1 pKa = 6.33TTWTIQGFSGSDD13 pKa = 3.29LIIAEE18 pKa = 4.58GGSTLSVGSNFMLDD32 pKa = 3.84PTWDD36 pKa = 3.35ASTDD40 pKa = 3.24ATSYY44 pKa = 10.63TFTDD48 pKa = 3.79PDD50 pKa = 3.75DD51 pKa = 4.25TNLSGDD57 pKa = 3.75NLNNEE62 pKa = 4.64HH63 pKa = 6.9GNDD66 pKa = 3.68GNQTINVYY74 pKa = 10.42DD75 pKa = 3.48SSGTLIHH82 pKa = 6.61SGQVYY87 pKa = 10.17IEE89 pKa = 4.64DD90 pKa = 3.75SAVFTAPDD98 pKa = 3.86GSSITLYY105 pKa = 9.16TLEE108 pKa = 4.14VNGVVVGTVTSAPLVPGVTYY128 pKa = 10.47HH129 pKa = 5.02VTAINDD135 pKa = 3.48VTTGPLYY142 pKa = 10.16TALASTTYY150 pKa = 11.08DD151 pKa = 3.42PDD153 pKa = 3.47LANTVTGGDD162 pKa = 3.71YY163 pKa = 11.07ADD165 pKa = 5.22SITGGAGNDD174 pKa = 4.08TITTGAGSDD183 pKa = 3.88TIDD186 pKa = 3.66GGTGNDD192 pKa = 3.7SIQFGTGGDD201 pKa = 3.87SVHH204 pKa = 6.93GGDD207 pKa = 4.03GNDD210 pKa = 4.42YY211 pKa = 10.44IDD213 pKa = 5.72DD214 pKa = 4.15YY215 pKa = 11.62AGSTGYY221 pKa = 10.74VYY223 pKa = 11.24NDD225 pKa = 3.72TLYY228 pKa = 11.47GDD230 pKa = 4.74AGNDD234 pKa = 3.62TIFAGSGNDD243 pKa = 3.77YY244 pKa = 11.27VDD246 pKa = 4.45GGADD250 pKa = 3.21NDD252 pKa = 4.06NIYY255 pKa = 11.21GEE257 pKa = 5.03DD258 pKa = 4.07GNDD261 pKa = 3.51TLLGGAGCDD270 pKa = 3.6WIEE273 pKa = 4.35GGAGADD279 pKa = 4.12SIDD282 pKa = 3.92GGTGTDD288 pKa = 3.86IILGGDD294 pKa = 3.75GNDD297 pKa = 4.2TIHH300 pKa = 6.85GGDD303 pKa = 3.56GADD306 pKa = 3.37YY307 pKa = 10.94LAGNTGNDD315 pKa = 3.35VLYY318 pKa = 11.21GEE320 pKa = 5.24ADD322 pKa = 3.17ADD324 pKa = 3.54RR325 pKa = 11.84FYY327 pKa = 11.43FEE329 pKa = 5.42NGWGSDD335 pKa = 3.21TVYY338 pKa = 11.0GGSTTTAGGVDD349 pKa = 5.12LDD351 pKa = 4.4TLDD354 pKa = 4.06FSYY357 pKa = 9.84DD358 pKa = 3.48TVSGVSVTFTDD369 pKa = 3.8WEE371 pKa = 4.95DD372 pKa = 3.38GTATDD377 pKa = 4.0GTSTVTFDD385 pKa = 4.25NIEE388 pKa = 4.51GIYY391 pKa = 9.1GTAFGDD397 pKa = 4.06TINASADD404 pKa = 3.51GSGLTLDD411 pKa = 4.2GQAGNDD417 pKa = 3.94SIIGGSGDD425 pKa = 3.75DD426 pKa = 4.74CILGGAGDD434 pKa = 4.0DD435 pKa = 4.23TIFSGFGNDD444 pKa = 3.01TVYY447 pKa = 11.14GGDD450 pKa = 3.89GNDD453 pKa = 4.59SIDD456 pKa = 4.07DD457 pKa = 3.75KK458 pKa = 11.64SGVLAEE464 pKa = 4.52TYY466 pKa = 10.07TNYY469 pKa = 10.09VDD471 pKa = 5.24AGAGNDD477 pKa = 3.64TVFTGGGSDD486 pKa = 3.71TILGGEE492 pKa = 4.92GDD494 pKa = 4.34DD495 pKa = 4.3QLHH498 pKa = 6.64AEE500 pKa = 4.52AGDD503 pKa = 3.63DD504 pKa = 3.99LVYY507 pKa = 11.03GGAGADD513 pKa = 3.63TLWGNEE519 pKa = 4.11GADD522 pKa = 3.97TLWGGDD528 pKa = 3.5GADD531 pKa = 4.53SILGGIGDD539 pKa = 4.05DD540 pKa = 5.08LIYY543 pKa = 11.25GEE545 pKa = 5.27AGNDD549 pKa = 3.82TITGGDD555 pKa = 3.62GADD558 pKa = 4.16SISGGADD565 pKa = 2.9NDD567 pKa = 4.27VITGDD572 pKa = 4.74LGDD575 pKa = 4.26DD576 pKa = 3.71TLAGGTGADD585 pKa = 3.73TLTGGTGADD594 pKa = 3.59TYY596 pKa = 11.79VMSDD600 pKa = 2.87GDD602 pKa = 4.01GADD605 pKa = 3.24TVTDD609 pKa = 3.91FDD611 pKa = 3.72MTMVSGLTTDD621 pKa = 3.29QFDD624 pKa = 3.74VSGLKK629 pKa = 10.18DD630 pKa = 3.42AEE632 pKa = 4.28GNPVNVWDD640 pKa = 3.92VTVDD644 pKa = 3.33SDD646 pKa = 3.85EE647 pKa = 4.14QGNAVLFFPDD657 pKa = 3.75GTSVTLIGVSKK668 pKa = 9.65TSVNTAPMLHH678 pKa = 6.68AMGIPCLVAGSPVATPRR695 pKa = 11.84GPVAVEE701 pKa = 3.42QLVPGDD707 pKa = 3.93LVITASGAAEE717 pKa = 3.74PVLWTGARR725 pKa = 11.84AVTAAEE731 pKa = 4.03MAADD735 pKa = 3.88PRR737 pKa = 11.84LLPVEE742 pKa = 4.51IGAGRR747 pKa = 11.84LGNAAPVRR755 pKa = 11.84LSALHH760 pKa = 6.12AVHH763 pKa = 6.47VPARR767 pKa = 11.84GGALARR773 pKa = 11.84AGHH776 pKa = 5.87MAATGWGGARR786 pKa = 11.84ILHH789 pKa = 5.67GLARR793 pKa = 11.84QEE795 pKa = 3.63AGLRR799 pKa = 11.84YY800 pKa = 9.25HH801 pKa = 6.89HH802 pKa = 7.04LLLPRR807 pKa = 11.84HH808 pKa = 5.83ALISVAGLWVEE819 pKa = 4.26SFWPGRR825 pKa = 11.84QGLAGLDD832 pKa = 3.5PASRR836 pKa = 11.84TRR838 pKa = 11.84LIRR841 pKa = 11.84ARR843 pKa = 11.84PQLAQVLWGGASVAACYY860 pKa = 9.04SQPAAPFLRR869 pKa = 11.84RR870 pKa = 11.84RR871 pKa = 11.84EE872 pKa = 3.9IDD874 pKa = 3.38RR875 pKa = 11.84AACSNWSLLARR886 pKa = 11.84EE887 pKa = 4.9KK888 pKa = 10.36PHH890 pKa = 7.43SDD892 pKa = 2.79GFQASSGQASTLL904 pKa = 3.57
Molecular weight: 91.34 kDa
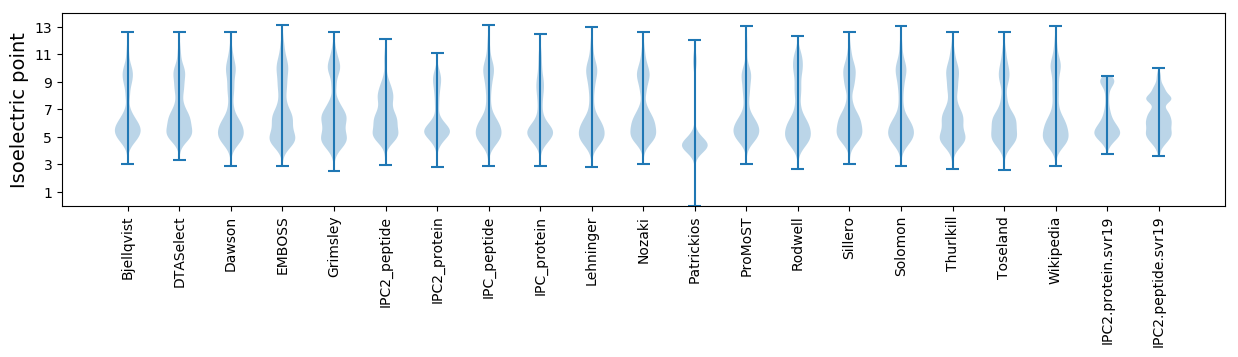
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A285RL35|A0A285RL35_9RHOB SSU ribosomal protein S13P OS=Rhodobacter maris OX=446682 GN=SAMN05877831_101642 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.43LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.38GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.43GGRR28 pKa = 11.84KK29 pKa = 8.99VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.53GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.43LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.38GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.43GGRR28 pKa = 11.84KK29 pKa = 8.99VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.53GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1149499 |
25 |
2405 |
311.8 |
33.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.953 ± 0.073 | 0.906 ± 0.013 |
5.335 ± 0.043 | 6.137 ± 0.041 |
3.602 ± 0.025 | 8.843 ± 0.044 |
1.98 ± 0.019 | 4.807 ± 0.027 |
3.031 ± 0.033 | 10.565 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.657 ± 0.02 | 2.192 ± 0.022 |
5.431 ± 0.038 | 2.921 ± 0.024 |
7.078 ± 0.047 | 4.801 ± 0.027 |
5.28 ± 0.029 | 7.089 ± 0.033 |
1.361 ± 0.016 | 2.031 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
