
Flammeovirgaceae bacterium 311
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Flammeovirgaceae; unclassified Flammeovirgaceae
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
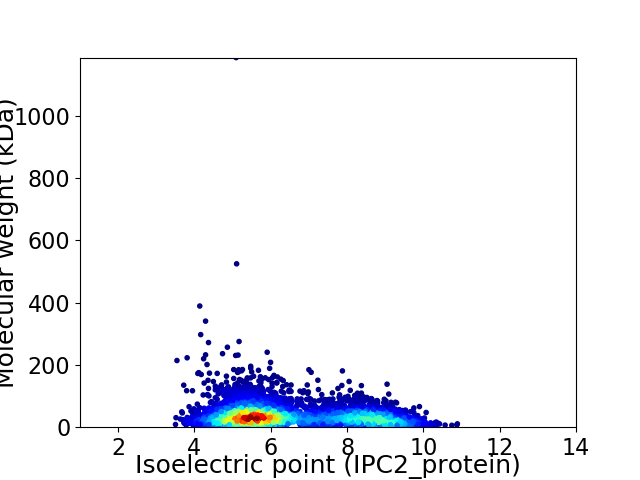
Virtual 2D-PAGE plot for 5332 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D3LIH2|A0A0D3LIH2_9BACT 4a-hydroxytetrahydrobiopterin dehydratase OS=Flammeovirgaceae bacterium 311 OX=1257021 GN=D770_17335 PE=3 SV=1
MM1 pKa = 7.52KK2 pKa = 10.49KK3 pKa = 8.67STLKK7 pKa = 10.65KK8 pKa = 9.01YY9 pKa = 9.59FLWSTLLLYY18 pKa = 9.26FMWSSNLVKK27 pKa = 10.11ATPAEE32 pKa = 4.14AAPALIFTSPGYY44 pKa = 10.27SEE46 pKa = 4.74NNKK49 pKa = 10.04NSLAAEE55 pKa = 4.41DD56 pKa = 4.17ADD58 pKa = 3.9GDD60 pKa = 4.38GVEE63 pKa = 4.74DD64 pKa = 4.31TVDD67 pKa = 3.52NCPSNYY73 pKa = 9.83NPGQEE78 pKa = 5.21DD79 pKa = 3.66MDD81 pKa = 5.62GDD83 pKa = 4.73GIGDD87 pKa = 3.84ACDD90 pKa = 4.66DD91 pKa = 5.0DD92 pKa = 4.19MDD94 pKa = 6.38GDD96 pKa = 4.16GALNEE101 pKa = 4.7LDD103 pKa = 5.05CNPNDD108 pKa = 3.86PADD111 pKa = 3.79SGEE114 pKa = 4.0YY115 pKa = 9.55WFADD119 pKa = 3.3EE120 pKa = 5.93DD121 pKa = 4.54GDD123 pKa = 4.89GFGSQNEE130 pKa = 4.09ILACEE135 pKa = 4.0QPEE138 pKa = 4.9GYY140 pKa = 10.05VDD142 pKa = 5.69NYY144 pKa = 11.47DD145 pKa = 4.07DD146 pKa = 6.2CNDD149 pKa = 3.53NNPNINPEE157 pKa = 4.2AEE159 pKa = 4.36DD160 pKa = 4.37DD161 pKa = 4.29PYY163 pKa = 12.11DD164 pKa = 5.12NIDD167 pKa = 4.06SNCDD171 pKa = 3.38GIDD174 pKa = 4.26PIGSDD179 pKa = 3.68ADD181 pKa = 3.96LDD183 pKa = 4.36GVDD186 pKa = 5.62DD187 pKa = 6.37LYY189 pKa = 11.68DD190 pKa = 4.06NCPDD194 pKa = 3.53TYY196 pKa = 11.27NPKK199 pKa = 10.0QEE201 pKa = 4.32DD202 pKa = 3.99TNEE205 pKa = 4.07DD206 pKa = 3.98GIGDD210 pKa = 3.81VCDD213 pKa = 5.11DD214 pKa = 4.88DD215 pKa = 4.72VAEE218 pKa = 4.35NQAPVITILDD228 pKa = 3.98YY229 pKa = 11.34DD230 pKa = 3.86PEE232 pKa = 4.98KK233 pKa = 10.82YY234 pKa = 10.44YY235 pKa = 11.0YY236 pKa = 10.67AAGQEE241 pKa = 4.42HH242 pKa = 6.76VFTIQVTDD250 pKa = 3.88PDD252 pKa = 4.08RR253 pKa = 11.84LEE255 pKa = 5.1LLLLDD260 pKa = 4.44IEE262 pKa = 4.67YY263 pKa = 10.97NKK265 pKa = 10.43NAYY268 pKa = 9.21QPGEE272 pKa = 4.14LTFSPSLPVRR282 pKa = 11.84GVGSLQIEE290 pKa = 4.7VRR292 pKa = 11.84WKK294 pKa = 10.26PNMEE298 pKa = 4.21RR299 pKa = 11.84PQHH302 pKa = 5.55LQLDD306 pKa = 4.9IIAMDD311 pKa = 4.14GDD313 pKa = 3.66RR314 pKa = 11.84EE315 pKa = 4.19RR316 pKa = 11.84TVKK319 pKa = 10.75YY320 pKa = 10.77LFGDD324 pKa = 4.25PYY326 pKa = 11.32LPTQISTPQEE336 pKa = 4.23PIVIGVDD343 pKa = 2.9QAFRR347 pKa = 11.84IDD349 pKa = 3.71IPFTADD355 pKa = 3.3FEE357 pKa = 4.57EE358 pKa = 4.66EE359 pKa = 3.96AYY361 pKa = 10.51AWFDD365 pKa = 3.62APEE368 pKa = 3.8WAADD372 pKa = 3.73DD373 pKa = 4.18NEE375 pKa = 4.41YY376 pKa = 9.35TLTLTNNEE384 pKa = 3.92FGMPAMTDD392 pKa = 2.76VGTYY396 pKa = 8.92QIPLGVSYY404 pKa = 9.66PWGTTRR410 pKa = 11.84STITVIVSAEE420 pKa = 3.95CTNQNWYY427 pKa = 9.11MDD429 pKa = 4.28EE430 pKa = 5.77DD431 pKa = 4.61GDD433 pKa = 5.35GYY435 pKa = 10.5GTQQTSAYY443 pKa = 9.84FPFRR447 pKa = 11.84QITCSSWPGYY457 pKa = 9.94VNNNDD462 pKa = 3.55DD463 pKa = 4.43CNDD466 pKa = 3.43NEE468 pKa = 4.84ASINPDD474 pKa = 2.97APEE477 pKa = 4.3IPGDD481 pKa = 4.87GIDD484 pKa = 3.93NNCNGQVDD492 pKa = 4.26DD493 pKa = 5.02GSTNTTAPYY502 pKa = 10.01FISSDD507 pKa = 2.91IGQEE511 pKa = 3.85ILVGGQFNFTIQVADD526 pKa = 3.62PTMGDD531 pKa = 3.5VVTLRR536 pKa = 11.84VDD538 pKa = 3.56SAAMINYY545 pKa = 8.46GSSGPPYY552 pKa = 10.3TLLPDD557 pKa = 3.98LAGATFSPDD566 pKa = 3.16RR567 pKa = 11.84SVTAADD573 pKa = 3.63TASLHH578 pKa = 5.83MNWTPTRR585 pKa = 11.84EE586 pKa = 4.22HH587 pKa = 6.9LGEE590 pKa = 4.04IYY592 pKa = 10.56FFFSATDD599 pKa = 3.17QHH601 pKa = 7.0GNSVWGFLPVVVDD614 pKa = 6.12LPFALQAVPPVVVGTQQPFEE634 pKa = 4.33VIIPISNSDD643 pKa = 2.89WHH645 pKa = 6.81QLDD648 pKa = 3.85EE649 pKa = 4.46VSLLEE654 pKa = 4.97GAPSWLQAALVSVDD668 pKa = 2.87GRR670 pKa = 11.84EE671 pKa = 3.91VPNAIRR677 pKa = 11.84VWGTPPARR685 pKa = 11.84SFGTYY690 pKa = 9.06KK691 pKa = 10.45AQVRR695 pKa = 11.84ARR697 pKa = 11.84TFWAKK702 pKa = 9.97WDD704 pKa = 3.8TLEE707 pKa = 4.4LQIRR711 pKa = 11.84VGNCTNRR718 pKa = 11.84AWYY721 pKa = 10.4ADD723 pKa = 3.46ADD725 pKa = 4.11GDD727 pKa = 5.17GFGADD732 pKa = 3.13NTMVTACWQPEE743 pKa = 4.68GYY745 pKa = 9.62VSKK748 pKa = 11.27GGDD751 pKa = 3.92CNDD754 pKa = 3.83ADD756 pKa = 3.44ATVYY760 pKa = 10.75PEE762 pKa = 4.35APEE765 pKa = 4.43LADD768 pKa = 5.23GKK770 pKa = 10.82DD771 pKa = 3.61NNCDD775 pKa = 3.19GMADD779 pKa = 3.59EE780 pKa = 5.48TDD782 pKa = 3.29NSCYY786 pKa = 8.64ATRR789 pKa = 11.84VISFTQGPRR798 pKa = 11.84ADD800 pKa = 3.42KK801 pKa = 10.81RR802 pKa = 11.84GVIDD806 pKa = 4.06PLRR809 pKa = 11.84SIPLRR814 pKa = 11.84ALGAPQEE821 pKa = 4.64DD822 pKa = 4.2KK823 pKa = 10.17THH825 pKa = 6.46SFVSLGFGGEE835 pKa = 4.16LVLEE839 pKa = 4.89LGANLYY845 pKa = 10.85DD846 pKa = 5.57DD847 pKa = 4.51GTTAPDD853 pKa = 3.48LMVVEE858 pKa = 5.0TTWGWANRR866 pKa = 11.84PCYY869 pKa = 9.76DD870 pKa = 3.27GKK872 pKa = 11.28GAGTLEE878 pKa = 3.99TMMMHH883 pKa = 6.34VSANGEE889 pKa = 3.88DD890 pKa = 3.91WVQVPGNFCRR900 pKa = 11.84NVKK903 pKa = 10.03VDD905 pKa = 3.4ISPVTGKK912 pKa = 10.67DD913 pKa = 2.88MLPYY917 pKa = 10.19VRR919 pKa = 11.84YY920 pKa = 9.95IKK922 pKa = 9.68ITDD925 pKa = 3.47TSDD928 pKa = 3.18PANFNRR934 pKa = 11.84SGNGYY939 pKa = 9.97DD940 pKa = 3.04VDD942 pKa = 5.16GIITCRR948 pKa = 11.84EE949 pKa = 4.14LFEE952 pKa = 4.83EE953 pKa = 4.73MPTNSRR959 pKa = 11.84TTAKK963 pKa = 10.65GFDD966 pKa = 3.42PNFFYY971 pKa = 10.67EE972 pKa = 4.05ALEE975 pKa = 4.33DD976 pKa = 4.26EE977 pKa = 4.79EE978 pKa = 5.82MSMQPLSFYY987 pKa = 10.08PNPVKK992 pKa = 10.62DD993 pKa = 3.61VLTIQTSSFGDD1004 pKa = 3.22EE1005 pKa = 4.07SLQVEE1010 pKa = 5.28VYY1012 pKa = 10.21SVAGVLVYY1020 pKa = 10.26KK1021 pKa = 10.29AAHH1024 pKa = 6.36RR1025 pKa = 11.84ADD1027 pKa = 3.45MHH1029 pKa = 7.59SGEE1032 pKa = 4.6LQVDD1036 pKa = 4.53LQQLRR1041 pKa = 11.84QGVYY1045 pKa = 10.45LLRR1048 pKa = 11.84LQGEE1052 pKa = 4.61YY1053 pKa = 9.74QQQTLKK1059 pKa = 10.44ISKK1062 pKa = 9.31EE1063 pKa = 3.91
MM1 pKa = 7.52KK2 pKa = 10.49KK3 pKa = 8.67STLKK7 pKa = 10.65KK8 pKa = 9.01YY9 pKa = 9.59FLWSTLLLYY18 pKa = 9.26FMWSSNLVKK27 pKa = 10.11ATPAEE32 pKa = 4.14AAPALIFTSPGYY44 pKa = 10.27SEE46 pKa = 4.74NNKK49 pKa = 10.04NSLAAEE55 pKa = 4.41DD56 pKa = 4.17ADD58 pKa = 3.9GDD60 pKa = 4.38GVEE63 pKa = 4.74DD64 pKa = 4.31TVDD67 pKa = 3.52NCPSNYY73 pKa = 9.83NPGQEE78 pKa = 5.21DD79 pKa = 3.66MDD81 pKa = 5.62GDD83 pKa = 4.73GIGDD87 pKa = 3.84ACDD90 pKa = 4.66DD91 pKa = 5.0DD92 pKa = 4.19MDD94 pKa = 6.38GDD96 pKa = 4.16GALNEE101 pKa = 4.7LDD103 pKa = 5.05CNPNDD108 pKa = 3.86PADD111 pKa = 3.79SGEE114 pKa = 4.0YY115 pKa = 9.55WFADD119 pKa = 3.3EE120 pKa = 5.93DD121 pKa = 4.54GDD123 pKa = 4.89GFGSQNEE130 pKa = 4.09ILACEE135 pKa = 4.0QPEE138 pKa = 4.9GYY140 pKa = 10.05VDD142 pKa = 5.69NYY144 pKa = 11.47DD145 pKa = 4.07DD146 pKa = 6.2CNDD149 pKa = 3.53NNPNINPEE157 pKa = 4.2AEE159 pKa = 4.36DD160 pKa = 4.37DD161 pKa = 4.29PYY163 pKa = 12.11DD164 pKa = 5.12NIDD167 pKa = 4.06SNCDD171 pKa = 3.38GIDD174 pKa = 4.26PIGSDD179 pKa = 3.68ADD181 pKa = 3.96LDD183 pKa = 4.36GVDD186 pKa = 5.62DD187 pKa = 6.37LYY189 pKa = 11.68DD190 pKa = 4.06NCPDD194 pKa = 3.53TYY196 pKa = 11.27NPKK199 pKa = 10.0QEE201 pKa = 4.32DD202 pKa = 3.99TNEE205 pKa = 4.07DD206 pKa = 3.98GIGDD210 pKa = 3.81VCDD213 pKa = 5.11DD214 pKa = 4.88DD215 pKa = 4.72VAEE218 pKa = 4.35NQAPVITILDD228 pKa = 3.98YY229 pKa = 11.34DD230 pKa = 3.86PEE232 pKa = 4.98KK233 pKa = 10.82YY234 pKa = 10.44YY235 pKa = 11.0YY236 pKa = 10.67AAGQEE241 pKa = 4.42HH242 pKa = 6.76VFTIQVTDD250 pKa = 3.88PDD252 pKa = 4.08RR253 pKa = 11.84LEE255 pKa = 5.1LLLLDD260 pKa = 4.44IEE262 pKa = 4.67YY263 pKa = 10.97NKK265 pKa = 10.43NAYY268 pKa = 9.21QPGEE272 pKa = 4.14LTFSPSLPVRR282 pKa = 11.84GVGSLQIEE290 pKa = 4.7VRR292 pKa = 11.84WKK294 pKa = 10.26PNMEE298 pKa = 4.21RR299 pKa = 11.84PQHH302 pKa = 5.55LQLDD306 pKa = 4.9IIAMDD311 pKa = 4.14GDD313 pKa = 3.66RR314 pKa = 11.84EE315 pKa = 4.19RR316 pKa = 11.84TVKK319 pKa = 10.75YY320 pKa = 10.77LFGDD324 pKa = 4.25PYY326 pKa = 11.32LPTQISTPQEE336 pKa = 4.23PIVIGVDD343 pKa = 2.9QAFRR347 pKa = 11.84IDD349 pKa = 3.71IPFTADD355 pKa = 3.3FEE357 pKa = 4.57EE358 pKa = 4.66EE359 pKa = 3.96AYY361 pKa = 10.51AWFDD365 pKa = 3.62APEE368 pKa = 3.8WAADD372 pKa = 3.73DD373 pKa = 4.18NEE375 pKa = 4.41YY376 pKa = 9.35TLTLTNNEE384 pKa = 3.92FGMPAMTDD392 pKa = 2.76VGTYY396 pKa = 8.92QIPLGVSYY404 pKa = 9.66PWGTTRR410 pKa = 11.84STITVIVSAEE420 pKa = 3.95CTNQNWYY427 pKa = 9.11MDD429 pKa = 4.28EE430 pKa = 5.77DD431 pKa = 4.61GDD433 pKa = 5.35GYY435 pKa = 10.5GTQQTSAYY443 pKa = 9.84FPFRR447 pKa = 11.84QITCSSWPGYY457 pKa = 9.94VNNNDD462 pKa = 3.55DD463 pKa = 4.43CNDD466 pKa = 3.43NEE468 pKa = 4.84ASINPDD474 pKa = 2.97APEE477 pKa = 4.3IPGDD481 pKa = 4.87GIDD484 pKa = 3.93NNCNGQVDD492 pKa = 4.26DD493 pKa = 5.02GSTNTTAPYY502 pKa = 10.01FISSDD507 pKa = 2.91IGQEE511 pKa = 3.85ILVGGQFNFTIQVADD526 pKa = 3.62PTMGDD531 pKa = 3.5VVTLRR536 pKa = 11.84VDD538 pKa = 3.56SAAMINYY545 pKa = 8.46GSSGPPYY552 pKa = 10.3TLLPDD557 pKa = 3.98LAGATFSPDD566 pKa = 3.16RR567 pKa = 11.84SVTAADD573 pKa = 3.63TASLHH578 pKa = 5.83MNWTPTRR585 pKa = 11.84EE586 pKa = 4.22HH587 pKa = 6.9LGEE590 pKa = 4.04IYY592 pKa = 10.56FFFSATDD599 pKa = 3.17QHH601 pKa = 7.0GNSVWGFLPVVVDD614 pKa = 6.12LPFALQAVPPVVVGTQQPFEE634 pKa = 4.33VIIPISNSDD643 pKa = 2.89WHH645 pKa = 6.81QLDD648 pKa = 3.85EE649 pKa = 4.46VSLLEE654 pKa = 4.97GAPSWLQAALVSVDD668 pKa = 2.87GRR670 pKa = 11.84EE671 pKa = 3.91VPNAIRR677 pKa = 11.84VWGTPPARR685 pKa = 11.84SFGTYY690 pKa = 9.06KK691 pKa = 10.45AQVRR695 pKa = 11.84ARR697 pKa = 11.84TFWAKK702 pKa = 9.97WDD704 pKa = 3.8TLEE707 pKa = 4.4LQIRR711 pKa = 11.84VGNCTNRR718 pKa = 11.84AWYY721 pKa = 10.4ADD723 pKa = 3.46ADD725 pKa = 4.11GDD727 pKa = 5.17GFGADD732 pKa = 3.13NTMVTACWQPEE743 pKa = 4.68GYY745 pKa = 9.62VSKK748 pKa = 11.27GGDD751 pKa = 3.92CNDD754 pKa = 3.83ADD756 pKa = 3.44ATVYY760 pKa = 10.75PEE762 pKa = 4.35APEE765 pKa = 4.43LADD768 pKa = 5.23GKK770 pKa = 10.82DD771 pKa = 3.61NNCDD775 pKa = 3.19GMADD779 pKa = 3.59EE780 pKa = 5.48TDD782 pKa = 3.29NSCYY786 pKa = 8.64ATRR789 pKa = 11.84VISFTQGPRR798 pKa = 11.84ADD800 pKa = 3.42KK801 pKa = 10.81RR802 pKa = 11.84GVIDD806 pKa = 4.06PLRR809 pKa = 11.84SIPLRR814 pKa = 11.84ALGAPQEE821 pKa = 4.64DD822 pKa = 4.2KK823 pKa = 10.17THH825 pKa = 6.46SFVSLGFGGEE835 pKa = 4.16LVLEE839 pKa = 4.89LGANLYY845 pKa = 10.85DD846 pKa = 5.57DD847 pKa = 4.51GTTAPDD853 pKa = 3.48LMVVEE858 pKa = 5.0TTWGWANRR866 pKa = 11.84PCYY869 pKa = 9.76DD870 pKa = 3.27GKK872 pKa = 11.28GAGTLEE878 pKa = 3.99TMMMHH883 pKa = 6.34VSANGEE889 pKa = 3.88DD890 pKa = 3.91WVQVPGNFCRR900 pKa = 11.84NVKK903 pKa = 10.03VDD905 pKa = 3.4ISPVTGKK912 pKa = 10.67DD913 pKa = 2.88MLPYY917 pKa = 10.19VRR919 pKa = 11.84YY920 pKa = 9.95IKK922 pKa = 9.68ITDD925 pKa = 3.47TSDD928 pKa = 3.18PANFNRR934 pKa = 11.84SGNGYY939 pKa = 9.97DD940 pKa = 3.04VDD942 pKa = 5.16GIITCRR948 pKa = 11.84EE949 pKa = 4.14LFEE952 pKa = 4.83EE953 pKa = 4.73MPTNSRR959 pKa = 11.84TTAKK963 pKa = 10.65GFDD966 pKa = 3.42PNFFYY971 pKa = 10.67EE972 pKa = 4.05ALEE975 pKa = 4.33DD976 pKa = 4.26EE977 pKa = 4.79EE978 pKa = 5.82MSMQPLSFYY987 pKa = 10.08PNPVKK992 pKa = 10.62DD993 pKa = 3.61VLTIQTSSFGDD1004 pKa = 3.22EE1005 pKa = 4.07SLQVEE1010 pKa = 5.28VYY1012 pKa = 10.21SVAGVLVYY1020 pKa = 10.26KK1021 pKa = 10.29AAHH1024 pKa = 6.36RR1025 pKa = 11.84ADD1027 pKa = 3.45MHH1029 pKa = 7.59SGEE1032 pKa = 4.6LQVDD1036 pKa = 4.53LQQLRR1041 pKa = 11.84QGVYY1045 pKa = 10.45LLRR1048 pKa = 11.84LQGEE1052 pKa = 4.61YY1053 pKa = 9.74QQQTLKK1059 pKa = 10.44ISKK1062 pKa = 9.31EE1063 pKa = 3.91
Molecular weight: 117.09 kDa
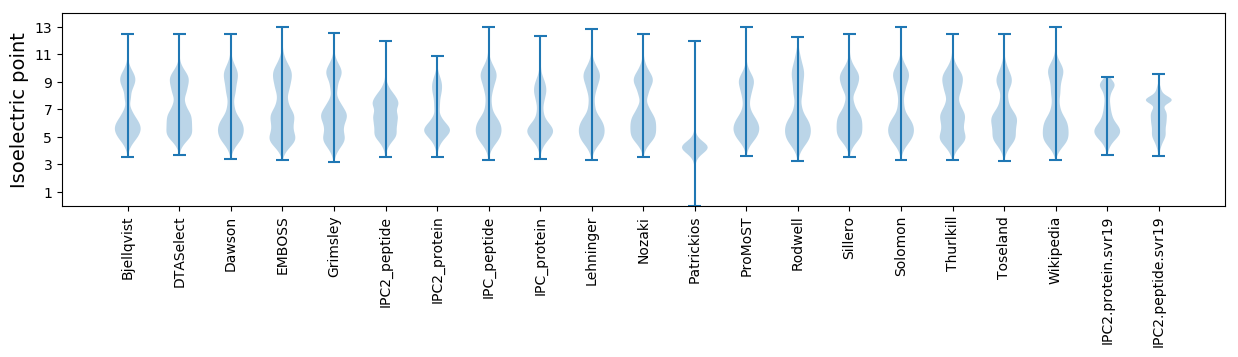
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D3LNQ4|A0A0D3LNQ4_9BACT Uncharacterized protein OS=Flammeovirgaceae bacterium 311 OX=1257021 GN=D770_26780 PE=4 SV=1
MM1 pKa = 7.59FFLQLLVFASPRR13 pKa = 11.84GPGGTTNQSQAGSPPKK29 pKa = 10.17RR30 pKa = 11.84GPPRR34 pKa = 11.84GNLVSPRR41 pKa = 11.84IEE43 pKa = 3.52ARR45 pKa = 11.84LRR47 pKa = 11.84QRR49 pKa = 11.84SRR51 pKa = 11.84RR52 pKa = 11.84GTATVALRR60 pKa = 11.84FAAFAKK66 pKa = 10.43AVLLINFSKK75 pKa = 10.71FRR77 pKa = 11.84MTHH80 pKa = 5.76FLVPYY85 pKa = 10.0LSKK88 pKa = 10.69FQGRR92 pKa = 11.84VKK94 pKa = 10.76GAFF97 pKa = 3.2
MM1 pKa = 7.59FFLQLLVFASPRR13 pKa = 11.84GPGGTTNQSQAGSPPKK29 pKa = 10.17RR30 pKa = 11.84GPPRR34 pKa = 11.84GNLVSPRR41 pKa = 11.84IEE43 pKa = 3.52ARR45 pKa = 11.84LRR47 pKa = 11.84QRR49 pKa = 11.84SRR51 pKa = 11.84RR52 pKa = 11.84GTATVALRR60 pKa = 11.84FAAFAKK66 pKa = 10.43AVLLINFSKK75 pKa = 10.71FRR77 pKa = 11.84MTHH80 pKa = 5.76FLVPYY85 pKa = 10.0LSKK88 pKa = 10.69FQGRR92 pKa = 11.84VKK94 pKa = 10.76GAFF97 pKa = 3.2
Molecular weight: 10.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1897602 |
33 |
10634 |
355.9 |
39.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.917 ± 0.03 | 0.743 ± 0.011 |
5.018 ± 0.026 | 6.606 ± 0.043 |
4.543 ± 0.027 | 7.077 ± 0.038 |
2.063 ± 0.014 | 6.242 ± 0.03 |
5.68 ± 0.037 | 10.235 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.309 ± 0.015 | 4.739 ± 0.033 |
4.177 ± 0.021 | 4.428 ± 0.03 |
4.777 ± 0.022 | 6.236 ± 0.036 |
5.351 ± 0.037 | 6.485 ± 0.027 |
1.34 ± 0.013 | 4.034 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
