
Wickerhamomyces anomalus (strain ATCC 58044 / CBS 1984 / NCYC 433 / NRRL Y-366-8) (Yeast) (Hansenula anomala)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Phaffomycetaceae; Wickerhamomyces; Wickerhamomyces anomalus
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
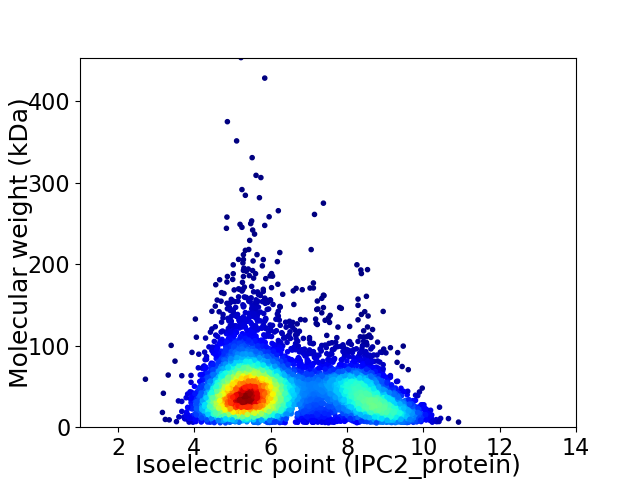
Virtual 2D-PAGE plot for 6406 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E3NYD3|A0A1E3NYD3_WICAA Uncharacterized protein OS=Wickerhamomyces anomalus (strain ATCC 58044 / CBS 1984 / NCYC 433 / NRRL Y-366-8) OX=683960 GN=WICANDRAFT_64249 PE=4 SV=1
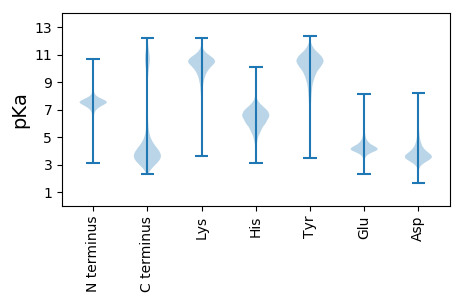
MM1 pKa = 8.0PSRR4 pKa = 11.84SYY6 pKa = 10.78ISPPSSTHH14 pKa = 6.11SDD16 pKa = 3.19SNEE19 pKa = 3.66PQNLVNPIVLPPIRR33 pKa = 11.84SLFDD37 pKa = 3.17EE38 pKa = 4.73GRR40 pKa = 11.84LPFFVDD46 pKa = 3.52QVEE49 pKa = 4.28DD50 pKa = 4.09LPFGGADD57 pKa = 3.97DD58 pKa = 4.24PVAPITTHH66 pKa = 5.9RR67 pKa = 11.84HH68 pKa = 5.44DD69 pKa = 4.71PGSAFSDD76 pKa = 3.54SSSDD80 pKa = 2.94ISSIFSQDD88 pKa = 3.06SAPISPPLTASLHH101 pKa = 5.26STASSDD107 pKa = 2.89HH108 pKa = 6.82FYY110 pKa = 11.14FEE112 pKa = 4.93EE113 pKa = 5.19LPLSPPPPYY122 pKa = 10.34SEE124 pKa = 5.53LEE126 pKa = 4.28SFNLNHH132 pKa = 6.58NFTSDD137 pKa = 3.75DD138 pKa = 3.52PFNFYY143 pKa = 10.64LVEE146 pKa = 4.06EE147 pKa = 4.47VAYY150 pKa = 9.58EE151 pKa = 3.9HH152 pKa = 7.25AYY154 pKa = 10.21EE155 pKa = 4.45LAQAAYY161 pKa = 10.65NSTDD165 pKa = 3.56LDD167 pKa = 4.08LDD169 pKa = 3.84SGVIADD175 pKa = 5.64DD176 pKa = 3.74EE177 pKa = 4.44MTNAGDD183 pKa = 3.44WDD185 pKa = 4.12YY186 pKa = 12.1LMTRR190 pKa = 11.84QTTAPEE196 pKa = 3.91TSNIMEE202 pKa = 4.67RR203 pKa = 11.84YY204 pKa = 7.28YY205 pKa = 11.21AGVRR209 pKa = 11.84FLQEE213 pKa = 3.97HH214 pKa = 6.71GLQPSEE220 pKa = 4.11IVSPTTFLYY229 pKa = 10.28SQNIEE234 pKa = 3.98NQYY237 pKa = 10.83LSTSIVSSEE246 pKa = 4.12QPSTPDD252 pKa = 3.37VSQHH256 pKa = 6.47PPLDD260 pKa = 4.11APSSEE265 pKa = 4.02THH267 pKa = 6.31YY268 pKa = 10.98EE269 pKa = 4.0DD270 pKa = 3.92PTFKK274 pKa = 9.93MDD276 pKa = 3.49TPIEE280 pKa = 4.0RR281 pKa = 11.84WIEE284 pKa = 3.9DD285 pKa = 3.14ASQEE289 pKa = 4.92DD290 pKa = 5.0EE291 pKa = 4.14VTEE294 pKa = 4.45DD295 pKa = 3.53NEE297 pKa = 4.91SVKK300 pKa = 10.32TPVNSPLVDD309 pKa = 4.28DD310 pKa = 4.36YY311 pKa = 12.11VDD313 pKa = 3.42YY314 pKa = 11.82DD315 pKa = 3.22MDD317 pKa = 3.64EE318 pKa = 4.39EE319 pKa = 5.05YY320 pKa = 10.97FFF322 pKa = 6.44
MM1 pKa = 8.0PSRR4 pKa = 11.84SYY6 pKa = 10.78ISPPSSTHH14 pKa = 6.11SDD16 pKa = 3.19SNEE19 pKa = 3.66PQNLVNPIVLPPIRR33 pKa = 11.84SLFDD37 pKa = 3.17EE38 pKa = 4.73GRR40 pKa = 11.84LPFFVDD46 pKa = 3.52QVEE49 pKa = 4.28DD50 pKa = 4.09LPFGGADD57 pKa = 3.97DD58 pKa = 4.24PVAPITTHH66 pKa = 5.9RR67 pKa = 11.84HH68 pKa = 5.44DD69 pKa = 4.71PGSAFSDD76 pKa = 3.54SSSDD80 pKa = 2.94ISSIFSQDD88 pKa = 3.06SAPISPPLTASLHH101 pKa = 5.26STASSDD107 pKa = 2.89HH108 pKa = 6.82FYY110 pKa = 11.14FEE112 pKa = 4.93EE113 pKa = 5.19LPLSPPPPYY122 pKa = 10.34SEE124 pKa = 5.53LEE126 pKa = 4.28SFNLNHH132 pKa = 6.58NFTSDD137 pKa = 3.75DD138 pKa = 3.52PFNFYY143 pKa = 10.64LVEE146 pKa = 4.06EE147 pKa = 4.47VAYY150 pKa = 9.58EE151 pKa = 3.9HH152 pKa = 7.25AYY154 pKa = 10.21EE155 pKa = 4.45LAQAAYY161 pKa = 10.65NSTDD165 pKa = 3.56LDD167 pKa = 4.08LDD169 pKa = 3.84SGVIADD175 pKa = 5.64DD176 pKa = 3.74EE177 pKa = 4.44MTNAGDD183 pKa = 3.44WDD185 pKa = 4.12YY186 pKa = 12.1LMTRR190 pKa = 11.84QTTAPEE196 pKa = 3.91TSNIMEE202 pKa = 4.67RR203 pKa = 11.84YY204 pKa = 7.28YY205 pKa = 11.21AGVRR209 pKa = 11.84FLQEE213 pKa = 3.97HH214 pKa = 6.71GLQPSEE220 pKa = 4.11IVSPTTFLYY229 pKa = 10.28SQNIEE234 pKa = 3.98NQYY237 pKa = 10.83LSTSIVSSEE246 pKa = 4.12QPSTPDD252 pKa = 3.37VSQHH256 pKa = 6.47PPLDD260 pKa = 4.11APSSEE265 pKa = 4.02THH267 pKa = 6.31YY268 pKa = 10.98EE269 pKa = 4.0DD270 pKa = 3.92PTFKK274 pKa = 9.93MDD276 pKa = 3.49TPIEE280 pKa = 4.0RR281 pKa = 11.84WIEE284 pKa = 3.9DD285 pKa = 3.14ASQEE289 pKa = 4.92DD290 pKa = 5.0EE291 pKa = 4.14VTEE294 pKa = 4.45DD295 pKa = 3.53NEE297 pKa = 4.91SVKK300 pKa = 10.32TPVNSPLVDD309 pKa = 4.28DD310 pKa = 4.36YY311 pKa = 12.11VDD313 pKa = 3.42YY314 pKa = 11.82DD315 pKa = 3.22MDD317 pKa = 3.64EE318 pKa = 4.39EE319 pKa = 5.05YY320 pKa = 10.97FFF322 pKa = 6.44
Molecular weight: 36.1 kDa
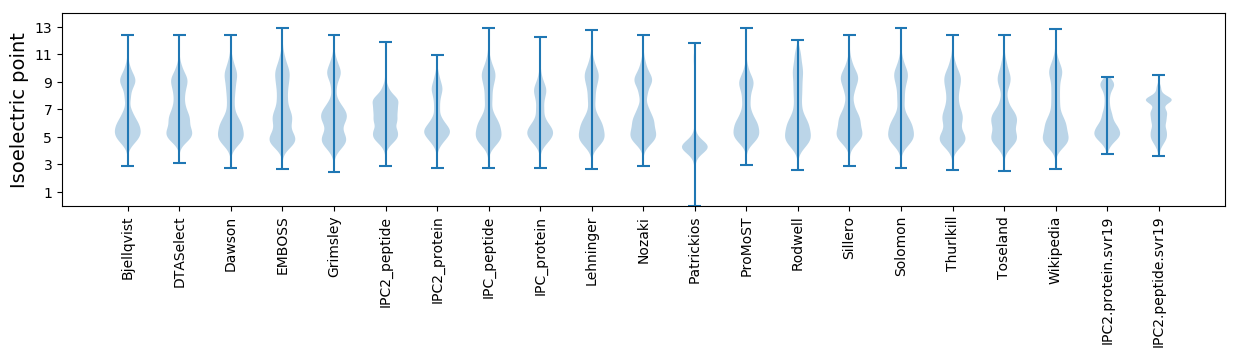
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E3P1N4|A0A1E3P1N4_WICAA Zn(2)-C6 fungal-type domain-containing protein OS=Wickerhamomyces anomalus (strain ATCC 58044 / CBS 1984 / NCYC 433 / NRRL Y-366-8) OX=683960 GN=WICANDRAFT_79636 PE=4 SV=1
NN1 pKa = 7.52SSTSSSTLLGMTPEE15 pKa = 3.92TARR18 pKa = 11.84RR19 pKa = 11.84NRR21 pKa = 11.84CTICQKK27 pKa = 8.75QFKK30 pKa = 9.67RR31 pKa = 11.84PSSLQTHH38 pKa = 7.29LYY40 pKa = 9.4SHH42 pKa = 6.85TGEE45 pKa = 4.61KK46 pKa = 10.34PFACTWDD53 pKa = 3.14NCGRR57 pKa = 11.84LFSVRR62 pKa = 11.84SNMIRR67 pKa = 11.84HH68 pKa = 6.14RR69 pKa = 11.84KK70 pKa = 7.35LHH72 pKa = 5.34EE73 pKa = 3.82RR74 pKa = 3.72
NN1 pKa = 7.52SSTSSSTLLGMTPEE15 pKa = 3.92TARR18 pKa = 11.84RR19 pKa = 11.84NRR21 pKa = 11.84CTICQKK27 pKa = 8.75QFKK30 pKa = 9.67RR31 pKa = 11.84PSSLQTHH38 pKa = 7.29LYY40 pKa = 9.4SHH42 pKa = 6.85TGEE45 pKa = 4.61KK46 pKa = 10.34PFACTWDD53 pKa = 3.14NCGRR57 pKa = 11.84LFSVRR62 pKa = 11.84SNMIRR67 pKa = 11.84HH68 pKa = 6.14RR69 pKa = 11.84KK70 pKa = 7.35LHH72 pKa = 5.34EE73 pKa = 3.82RR74 pKa = 3.72
Molecular weight: 8.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2782713 |
49 |
3951 |
434.4 |
49.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.561 ± 0.032 | 0.988 ± 0.011 |
5.988 ± 0.025 | 6.751 ± 0.029 |
4.768 ± 0.022 | 5.37 ± 0.03 |
2.048 ± 0.011 | 6.983 ± 0.028 |
7.446 ± 0.03 | 9.685 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.634 ± 0.011 | 5.773 ± 0.03 |
4.359 ± 0.028 | 4.062 ± 0.028 |
4.008 ± 0.02 | 8.628 ± 0.045 |
5.607 ± 0.02 | 5.818 ± 0.021 |
1.059 ± 0.009 | 3.465 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
