
Tenacibaculum sp. SG-28
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Tenacibaculum; unclassified Tenacibaculum
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
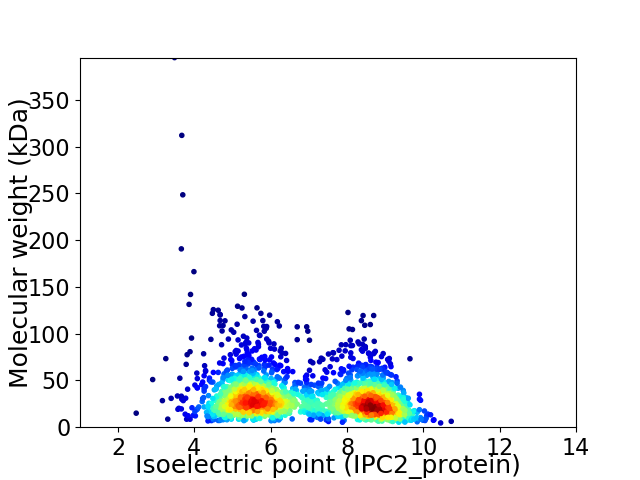
Virtual 2D-PAGE plot for 2136 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S7TMX1|A0A2S7TMX1_9FLAO Probable transcriptional regulatory protein BSU00_02810 OS=Tenacibaculum sp. SG-28 OX=754426 GN=BSU00_02810 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 10.19PEE4 pKa = 4.81LKK6 pKa = 10.6FSDD9 pKa = 3.58ACSSNGNRR17 pKa = 11.84DD18 pKa = 3.32FQIGFRR24 pKa = 11.84YY25 pKa = 7.72NTSTFQDD32 pKa = 3.64DD33 pKa = 4.37NIFTLEE39 pKa = 4.16LSDD42 pKa = 4.93ASGSFANPVNLATVANKK59 pKa = 9.9NATFSEE65 pKa = 4.18ILVAFQLPDD74 pKa = 3.3NTNGSGYY81 pKa = 10.3KK82 pKa = 9.76IRR84 pKa = 11.84IKK86 pKa = 10.52ADD88 pKa = 3.1KK89 pKa = 10.04PRR91 pKa = 11.84TSSQEE96 pKa = 3.9MFSPASDD103 pKa = 3.3AFAAYY108 pKa = 10.67LLFSEE113 pKa = 4.88TLWINDD119 pKa = 3.13NKK121 pKa = 10.78ASEE124 pKa = 4.66TICGSGSVRR133 pKa = 11.84LTLNTDD139 pKa = 3.09EE140 pKa = 4.67TASFDD145 pKa = 3.39WFKK148 pKa = 11.5DD149 pKa = 2.78GDD151 pKa = 4.03FFATTTEE158 pKa = 3.88PFIEE162 pKa = 4.29VSEE165 pKa = 4.03QGKK168 pKa = 8.83YY169 pKa = 7.92QGKK172 pKa = 9.78INYY175 pKa = 7.93FEE177 pKa = 4.64CGYY180 pKa = 10.61VDD182 pKa = 3.57SRR184 pKa = 11.84LIDD187 pKa = 3.59VYY189 pKa = 11.32VVSTTDD195 pKa = 3.38AAIQGGNEE203 pKa = 3.77VEE205 pKa = 4.11ICSDD209 pKa = 3.06EE210 pKa = 4.5TYY212 pKa = 10.39TFEE215 pKa = 5.71ASISDD220 pKa = 3.32TSYY223 pKa = 10.12TYY225 pKa = 11.0NWYY228 pKa = 10.59LDD230 pKa = 3.84DD231 pKa = 4.48QLVASSNSSTYY242 pKa = 7.35TTPNTGQFGIYY253 pKa = 10.19YY254 pKa = 10.28LILEE258 pKa = 4.68KK259 pKa = 10.73DD260 pKa = 3.17GCEE263 pKa = 3.93AKK265 pKa = 10.81SQDD268 pKa = 3.7VEE270 pKa = 4.79LKK272 pKa = 10.52QKK274 pKa = 9.22TDD276 pKa = 3.18SAIEE280 pKa = 4.48EE281 pKa = 4.1ITGDD285 pKa = 3.43FGTFLLLPGEE295 pKa = 4.2NKK297 pKa = 10.22KK298 pKa = 11.04LEE300 pKa = 3.94ISIAPSSVSTDD311 pKa = 3.54IIWLKK316 pKa = 10.92NDD318 pKa = 2.88VVLQGVSGLSMTPNGPGIYY337 pKa = 9.47KK338 pKa = 10.48ARR340 pKa = 11.84VTEE343 pKa = 4.39TGTSCPFTQDD353 pKa = 2.2SDD355 pKa = 4.01VFEE358 pKa = 5.2LVDD361 pKa = 4.43LQSLSPSIRR370 pKa = 11.84TASDD374 pKa = 3.56YY375 pKa = 10.11VACGIDD381 pKa = 3.43QTRR384 pKa = 11.84LLMVGITATGTDD396 pKa = 3.38GNTYY400 pKa = 9.95EE401 pKa = 5.79LSDD404 pKa = 3.86TQVNDD409 pKa = 3.14YY410 pKa = 10.69VQYY413 pKa = 10.56QWYY416 pKa = 9.84KK417 pKa = 9.7GGNIINGATQMEE429 pKa = 4.56FAIASYY435 pKa = 10.37QEE437 pKa = 3.87NDD439 pKa = 3.19SYY441 pKa = 11.25TLEE444 pKa = 4.18VSSGTFSGVSDD455 pKa = 5.36AIDD458 pKa = 3.02IQVTIEE464 pKa = 4.04SPEE467 pKa = 3.93ILSSSTSNALCTDD480 pKa = 3.58GTIVYY485 pKa = 8.9TIEE488 pKa = 3.92EE489 pKa = 4.34VVADD493 pKa = 4.92FTYY496 pKa = 9.65TWFKK500 pKa = 10.89DD501 pKa = 3.45GEE503 pKa = 4.45EE504 pKa = 4.5IEE506 pKa = 4.41TDD508 pKa = 3.29TPEE511 pKa = 3.89TLEE514 pKa = 4.2VIEE517 pKa = 4.42EE518 pKa = 4.31GVYY521 pKa = 10.17FLQISGFGCVTDD533 pKa = 4.65LEE535 pKa = 5.15SITVVPFDD543 pKa = 4.28DD544 pKa = 3.94SAVEE548 pKa = 4.04VTPSEE553 pKa = 4.47VVVVLPGQNVTVTASGANTYY573 pKa = 7.81EE574 pKa = 3.94WYY576 pKa = 9.84EE577 pKa = 4.2DD578 pKa = 3.48SSGSLLSTNEE588 pKa = 3.88TLDD591 pKa = 3.27INTVGTYY598 pKa = 8.11TLIATVDD605 pKa = 3.57DD606 pKa = 4.35CEE608 pKa = 4.35VIKK611 pKa = 10.03TIEE614 pKa = 4.14AVEE617 pKa = 4.01QDD619 pKa = 3.71DD620 pKa = 4.81QIIVPNIVSPYY631 pKa = 9.61LQDD634 pKa = 4.8GINDD638 pKa = 3.31TWTLSNKK645 pKa = 9.8YY646 pKa = 9.92AFQPSVTIAIYY657 pKa = 10.07DD658 pKa = 3.69ANGVEE663 pKa = 4.28VLRR666 pKa = 11.84TNEE669 pKa = 4.43YY670 pKa = 10.89KK671 pKa = 10.63NDD673 pKa = 3.61WPNKK677 pKa = 8.57DD678 pKa = 3.91LRR680 pKa = 11.84NQQVFYY686 pKa = 10.91YY687 pKa = 10.36KK688 pKa = 10.31IIRR691 pKa = 11.84DD692 pKa = 3.7DD693 pKa = 3.77MLIKK697 pKa = 10.59AGSISVLDD705 pKa = 3.78
MM1 pKa = 7.35KK2 pKa = 10.19PEE4 pKa = 4.81LKK6 pKa = 10.6FSDD9 pKa = 3.58ACSSNGNRR17 pKa = 11.84DD18 pKa = 3.32FQIGFRR24 pKa = 11.84YY25 pKa = 7.72NTSTFQDD32 pKa = 3.64DD33 pKa = 4.37NIFTLEE39 pKa = 4.16LSDD42 pKa = 4.93ASGSFANPVNLATVANKK59 pKa = 9.9NATFSEE65 pKa = 4.18ILVAFQLPDD74 pKa = 3.3NTNGSGYY81 pKa = 10.3KK82 pKa = 9.76IRR84 pKa = 11.84IKK86 pKa = 10.52ADD88 pKa = 3.1KK89 pKa = 10.04PRR91 pKa = 11.84TSSQEE96 pKa = 3.9MFSPASDD103 pKa = 3.3AFAAYY108 pKa = 10.67LLFSEE113 pKa = 4.88TLWINDD119 pKa = 3.13NKK121 pKa = 10.78ASEE124 pKa = 4.66TICGSGSVRR133 pKa = 11.84LTLNTDD139 pKa = 3.09EE140 pKa = 4.67TASFDD145 pKa = 3.39WFKK148 pKa = 11.5DD149 pKa = 2.78GDD151 pKa = 4.03FFATTTEE158 pKa = 3.88PFIEE162 pKa = 4.29VSEE165 pKa = 4.03QGKK168 pKa = 8.83YY169 pKa = 7.92QGKK172 pKa = 9.78INYY175 pKa = 7.93FEE177 pKa = 4.64CGYY180 pKa = 10.61VDD182 pKa = 3.57SRR184 pKa = 11.84LIDD187 pKa = 3.59VYY189 pKa = 11.32VVSTTDD195 pKa = 3.38AAIQGGNEE203 pKa = 3.77VEE205 pKa = 4.11ICSDD209 pKa = 3.06EE210 pKa = 4.5TYY212 pKa = 10.39TFEE215 pKa = 5.71ASISDD220 pKa = 3.32TSYY223 pKa = 10.12TYY225 pKa = 11.0NWYY228 pKa = 10.59LDD230 pKa = 3.84DD231 pKa = 4.48QLVASSNSSTYY242 pKa = 7.35TTPNTGQFGIYY253 pKa = 10.19YY254 pKa = 10.28LILEE258 pKa = 4.68KK259 pKa = 10.73DD260 pKa = 3.17GCEE263 pKa = 3.93AKK265 pKa = 10.81SQDD268 pKa = 3.7VEE270 pKa = 4.79LKK272 pKa = 10.52QKK274 pKa = 9.22TDD276 pKa = 3.18SAIEE280 pKa = 4.48EE281 pKa = 4.1ITGDD285 pKa = 3.43FGTFLLLPGEE295 pKa = 4.2NKK297 pKa = 10.22KK298 pKa = 11.04LEE300 pKa = 3.94ISIAPSSVSTDD311 pKa = 3.54IIWLKK316 pKa = 10.92NDD318 pKa = 2.88VVLQGVSGLSMTPNGPGIYY337 pKa = 9.47KK338 pKa = 10.48ARR340 pKa = 11.84VTEE343 pKa = 4.39TGTSCPFTQDD353 pKa = 2.2SDD355 pKa = 4.01VFEE358 pKa = 5.2LVDD361 pKa = 4.43LQSLSPSIRR370 pKa = 11.84TASDD374 pKa = 3.56YY375 pKa = 10.11VACGIDD381 pKa = 3.43QTRR384 pKa = 11.84LLMVGITATGTDD396 pKa = 3.38GNTYY400 pKa = 9.95EE401 pKa = 5.79LSDD404 pKa = 3.86TQVNDD409 pKa = 3.14YY410 pKa = 10.69VQYY413 pKa = 10.56QWYY416 pKa = 9.84KK417 pKa = 9.7GGNIINGATQMEE429 pKa = 4.56FAIASYY435 pKa = 10.37QEE437 pKa = 3.87NDD439 pKa = 3.19SYY441 pKa = 11.25TLEE444 pKa = 4.18VSSGTFSGVSDD455 pKa = 5.36AIDD458 pKa = 3.02IQVTIEE464 pKa = 4.04SPEE467 pKa = 3.93ILSSSTSNALCTDD480 pKa = 3.58GTIVYY485 pKa = 8.9TIEE488 pKa = 3.92EE489 pKa = 4.34VVADD493 pKa = 4.92FTYY496 pKa = 9.65TWFKK500 pKa = 10.89DD501 pKa = 3.45GEE503 pKa = 4.45EE504 pKa = 4.5IEE506 pKa = 4.41TDD508 pKa = 3.29TPEE511 pKa = 3.89TLEE514 pKa = 4.2VIEE517 pKa = 4.42EE518 pKa = 4.31GVYY521 pKa = 10.17FLQISGFGCVTDD533 pKa = 4.65LEE535 pKa = 5.15SITVVPFDD543 pKa = 4.28DD544 pKa = 3.94SAVEE548 pKa = 4.04VTPSEE553 pKa = 4.47VVVVLPGQNVTVTASGANTYY573 pKa = 7.81EE574 pKa = 3.94WYY576 pKa = 9.84EE577 pKa = 4.2DD578 pKa = 3.48SSGSLLSTNEE588 pKa = 3.88TLDD591 pKa = 3.27INTVGTYY598 pKa = 8.11TLIATVDD605 pKa = 3.57DD606 pKa = 4.35CEE608 pKa = 4.35VIKK611 pKa = 10.03TIEE614 pKa = 4.14AVEE617 pKa = 4.01QDD619 pKa = 3.71DD620 pKa = 4.81QIIVPNIVSPYY631 pKa = 9.61LQDD634 pKa = 4.8GINDD638 pKa = 3.31TWTLSNKK645 pKa = 9.8YY646 pKa = 9.92AFQPSVTIAIYY657 pKa = 10.07DD658 pKa = 3.69ANGVEE663 pKa = 4.28VLRR666 pKa = 11.84TNEE669 pKa = 4.43YY670 pKa = 10.89KK671 pKa = 10.63NDD673 pKa = 3.61WPNKK677 pKa = 8.57DD678 pKa = 3.91LRR680 pKa = 11.84NQQVFYY686 pKa = 10.91YY687 pKa = 10.36KK688 pKa = 10.31IIRR691 pKa = 11.84DD692 pKa = 3.7DD693 pKa = 3.77MLIKK697 pKa = 10.59AGSISVLDD705 pKa = 3.78
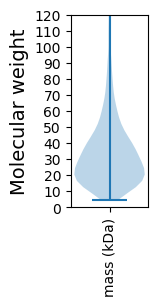
Molecular weight: 77.54 kDa
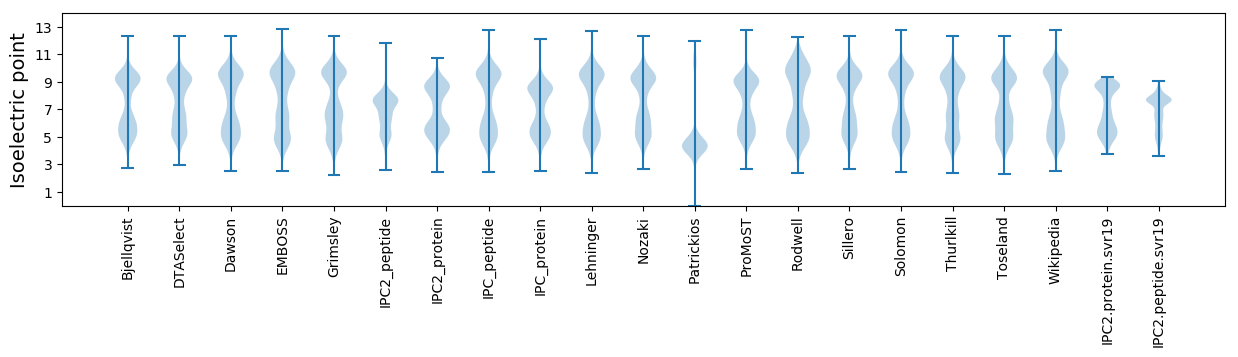
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S7TJL7|A0A2S7TJL7_9FLAO Peptide transporter OS=Tenacibaculum sp. SG-28 OX=754426 GN=BSU00_09150 PE=3 SV=1
MM1 pKa = 7.23SVQSVRR7 pKa = 11.84HH8 pKa = 4.4FFEE11 pKa = 5.25RR12 pKa = 11.84NGFNVSSRR20 pKa = 11.84LADD23 pKa = 3.79RR24 pKa = 11.84LGMRR28 pKa = 11.84AVNVRR33 pKa = 11.84LFFIYY38 pKa = 10.25ISFVTVGLSFGLYY51 pKa = 8.31LTLAFLLKK59 pKa = 10.31LKK61 pKa = 10.99DD62 pKa = 3.33MIYY65 pKa = 10.19TKK67 pKa = 9.83RR68 pKa = 11.84TSVFDD73 pKa = 3.66LL74 pKa = 4.18
MM1 pKa = 7.23SVQSVRR7 pKa = 11.84HH8 pKa = 4.4FFEE11 pKa = 5.25RR12 pKa = 11.84NGFNVSSRR20 pKa = 11.84LADD23 pKa = 3.79RR24 pKa = 11.84LGMRR28 pKa = 11.84AVNVRR33 pKa = 11.84LFFIYY38 pKa = 10.25ISFVTVGLSFGLYY51 pKa = 8.31LTLAFLLKK59 pKa = 10.31LKK61 pKa = 10.99DD62 pKa = 3.33MIYY65 pKa = 10.19TKK67 pKa = 9.83RR68 pKa = 11.84TSVFDD73 pKa = 3.66LL74 pKa = 4.18
Molecular weight: 8.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
658611 |
38 |
3746 |
308.3 |
34.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.503 ± 0.048 | 0.782 ± 0.017 |
5.289 ± 0.056 | 6.5 ± 0.053 |
5.187 ± 0.045 | 6.301 ± 0.058 |
1.766 ± 0.026 | 8.177 ± 0.046 |
8.129 ± 0.078 | 9.154 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.145 ± 0.026 | 6.154 ± 0.042 |
3.265 ± 0.026 | 3.365 ± 0.033 |
3.394 ± 0.038 | 6.487 ± 0.042 |
6.049 ± 0.065 | 6.313 ± 0.046 |
0.969 ± 0.017 | 4.071 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
