
Candidatus Bathyarchaeota archaeon
Taxonomy: cellular organisms; Archaea; TACK group; Candidatus Bathyarchaeota; unclassified Candidatus Bathyarchaeota
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
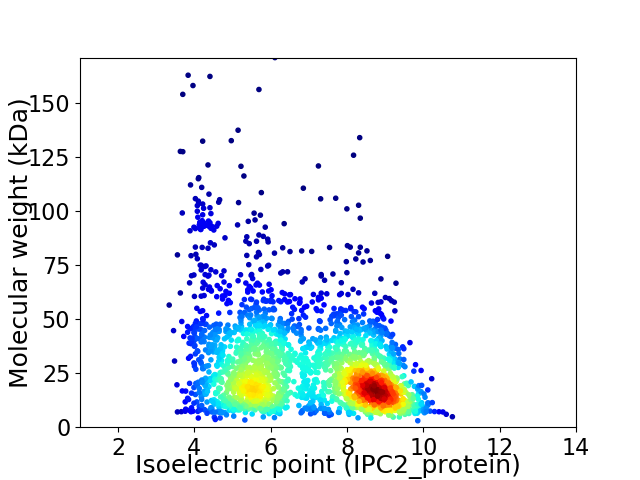
Virtual 2D-PAGE plot for 2329 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U0RXR1|A0A2U0RXR1_9ARCH Uncharacterized protein OS=Candidatus Bathyarchaeota archaeon OX=2026714 GN=CW691_09435 PE=4 SV=1
MM1 pKa = 7.46QINKK5 pKa = 9.61KK6 pKa = 10.3LSTGLIVTLLALSTIAIAIPSAFAVVANLQAYY38 pKa = 6.49TAMGAPITGGTVGQSIRR55 pKa = 11.84VGGDD59 pKa = 2.87SAAAWALVQFYY70 pKa = 10.02WDD72 pKa = 3.61SPANKK77 pKa = 9.1IGEE80 pKa = 4.35VYY82 pKa = 10.99ADD84 pKa = 3.61GAGDD88 pKa = 3.87FLLTGITVPEE98 pKa = 4.27DD99 pKa = 3.16VAGNHH104 pKa = 6.09QILVNDD110 pKa = 3.8GTGNVALNSNINPKK124 pKa = 8.99LTISATKK131 pKa = 10.22ALPGDD136 pKa = 3.94SLAVTGSGFGDD147 pKa = 3.75EE148 pKa = 4.66VPVGIYY154 pKa = 10.27LGTITSVVTEE164 pKa = 4.55AVADD168 pKa = 4.26DD169 pKa = 5.39DD170 pKa = 5.61LDD172 pKa = 3.79EE173 pKa = 5.23SPVVKK178 pKa = 9.62GTVVLTVDD186 pKa = 3.34VTVDD190 pKa = 3.18GDD192 pKa = 3.46IGGTAVTGPATLTVTDD208 pKa = 4.62DD209 pKa = 4.11GEE211 pKa = 4.73GVLSGTLSDD220 pKa = 3.89VVVDD224 pKa = 4.2DD225 pKa = 4.41GVVVEE230 pKa = 5.06DD231 pKa = 4.74GEE233 pKa = 4.43VDD235 pKa = 3.51VTIVGTINYY244 pKa = 7.16ATGGIALSASGVDD257 pKa = 3.64SAAGTGTVLNIAVTIDD273 pKa = 4.32AIDD276 pKa = 3.49AAYY279 pKa = 10.18QYY281 pKa = 11.11AQYY284 pKa = 10.85EE285 pKa = 4.38VTPLGGITTSAAGSFAEE302 pKa = 4.85SILVPAIPVLSYY314 pKa = 11.7GNYY317 pKa = 10.02LDD319 pKa = 4.17TAIDD323 pKa = 3.8TDD325 pKa = 4.28GNTANPTPVASLSVDD340 pKa = 3.24YY341 pKa = 10.8YY342 pKa = 10.76ILVNPISGPAGITVTLSGRR361 pKa = 11.84IPANTVYY368 pKa = 10.51EE369 pKa = 4.13IRR371 pKa = 11.84LDD373 pKa = 3.55TTTIATGTSAADD385 pKa = 3.28TTYY388 pKa = 10.0TQTHH392 pKa = 6.02QISSFLSKK400 pKa = 9.3TAHH403 pKa = 5.52TFYY406 pKa = 11.16VVWGGTNTRR415 pKa = 11.84SATFTVTEE423 pKa = 4.3SPRR426 pKa = 11.84MALSSTSGMAGQVITISSVPGYY448 pKa = 9.11PFSAGADD455 pKa = 3.22LTLYY459 pKa = 10.7INGAVVNGTTIDD471 pKa = 3.71DD472 pKa = 3.83RR473 pKa = 11.84FGPTVAFGPASGTFTDD489 pKa = 5.38LEE491 pKa = 4.26FTIPALAPGVYY502 pKa = 10.06AIEE505 pKa = 4.76LVDD508 pKa = 4.22SYY510 pKa = 11.67GASTGTMYY518 pKa = 10.73TFTVVPTPEE527 pKa = 4.19TAVSMVGTVFYY538 pKa = 10.63PGDD541 pKa = 3.43TLAWSFSSTDD551 pKa = 3.41TVTVAPTVTVRR562 pKa = 11.84SPTGAVWWQGVWATTTSGPMTTVLYY587 pKa = 10.6QNQLFGGAHH596 pKa = 5.79ATLPSNAPLGSWNWTAVYY614 pKa = 9.13STGSAANVKK623 pKa = 8.85ATGLFSVMAAATLDD637 pKa = 3.7TVTTDD642 pKa = 3.41IADD645 pKa = 4.05LVDD648 pKa = 5.54DD649 pKa = 5.02ISDD652 pKa = 3.45ISDD655 pKa = 3.53DD656 pKa = 3.61VATINTNINNLEE668 pKa = 4.07DD669 pKa = 5.57LIDD672 pKa = 4.09ALEE675 pKa = 4.23VTVDD679 pKa = 3.26IPEE682 pKa = 4.46LTTLTNDD689 pKa = 3.09VASLKK694 pKa = 10.54VSVSALDD701 pKa = 3.69AVVTAVAGDD710 pKa = 3.75VATVDD715 pKa = 3.76TKK717 pKa = 10.87IGTLTGTITGIEE729 pKa = 4.15GNIATIEE736 pKa = 4.17TDD738 pKa = 3.23VGTLQADD745 pKa = 3.48ISDD748 pKa = 3.82VEE750 pKa = 4.67ANVDD754 pKa = 3.73STPAWIAVVLALVAAVAAIFAVITIRR780 pKa = 11.84QKK782 pKa = 10.44IAGG785 pKa = 3.82
MM1 pKa = 7.46QINKK5 pKa = 9.61KK6 pKa = 10.3LSTGLIVTLLALSTIAIAIPSAFAVVANLQAYY38 pKa = 6.49TAMGAPITGGTVGQSIRR55 pKa = 11.84VGGDD59 pKa = 2.87SAAAWALVQFYY70 pKa = 10.02WDD72 pKa = 3.61SPANKK77 pKa = 9.1IGEE80 pKa = 4.35VYY82 pKa = 10.99ADD84 pKa = 3.61GAGDD88 pKa = 3.87FLLTGITVPEE98 pKa = 4.27DD99 pKa = 3.16VAGNHH104 pKa = 6.09QILVNDD110 pKa = 3.8GTGNVALNSNINPKK124 pKa = 8.99LTISATKK131 pKa = 10.22ALPGDD136 pKa = 3.94SLAVTGSGFGDD147 pKa = 3.75EE148 pKa = 4.66VPVGIYY154 pKa = 10.27LGTITSVVTEE164 pKa = 4.55AVADD168 pKa = 4.26DD169 pKa = 5.39DD170 pKa = 5.61LDD172 pKa = 3.79EE173 pKa = 5.23SPVVKK178 pKa = 9.62GTVVLTVDD186 pKa = 3.34VTVDD190 pKa = 3.18GDD192 pKa = 3.46IGGTAVTGPATLTVTDD208 pKa = 4.62DD209 pKa = 4.11GEE211 pKa = 4.73GVLSGTLSDD220 pKa = 3.89VVVDD224 pKa = 4.2DD225 pKa = 4.41GVVVEE230 pKa = 5.06DD231 pKa = 4.74GEE233 pKa = 4.43VDD235 pKa = 3.51VTIVGTINYY244 pKa = 7.16ATGGIALSASGVDD257 pKa = 3.64SAAGTGTVLNIAVTIDD273 pKa = 4.32AIDD276 pKa = 3.49AAYY279 pKa = 10.18QYY281 pKa = 11.11AQYY284 pKa = 10.85EE285 pKa = 4.38VTPLGGITTSAAGSFAEE302 pKa = 4.85SILVPAIPVLSYY314 pKa = 11.7GNYY317 pKa = 10.02LDD319 pKa = 4.17TAIDD323 pKa = 3.8TDD325 pKa = 4.28GNTANPTPVASLSVDD340 pKa = 3.24YY341 pKa = 10.8YY342 pKa = 10.76ILVNPISGPAGITVTLSGRR361 pKa = 11.84IPANTVYY368 pKa = 10.51EE369 pKa = 4.13IRR371 pKa = 11.84LDD373 pKa = 3.55TTTIATGTSAADD385 pKa = 3.28TTYY388 pKa = 10.0TQTHH392 pKa = 6.02QISSFLSKK400 pKa = 9.3TAHH403 pKa = 5.52TFYY406 pKa = 11.16VVWGGTNTRR415 pKa = 11.84SATFTVTEE423 pKa = 4.3SPRR426 pKa = 11.84MALSSTSGMAGQVITISSVPGYY448 pKa = 9.11PFSAGADD455 pKa = 3.22LTLYY459 pKa = 10.7INGAVVNGTTIDD471 pKa = 3.71DD472 pKa = 3.83RR473 pKa = 11.84FGPTVAFGPASGTFTDD489 pKa = 5.38LEE491 pKa = 4.26FTIPALAPGVYY502 pKa = 10.06AIEE505 pKa = 4.76LVDD508 pKa = 4.22SYY510 pKa = 11.67GASTGTMYY518 pKa = 10.73TFTVVPTPEE527 pKa = 4.19TAVSMVGTVFYY538 pKa = 10.63PGDD541 pKa = 3.43TLAWSFSSTDD551 pKa = 3.41TVTVAPTVTVRR562 pKa = 11.84SPTGAVWWQGVWATTTSGPMTTVLYY587 pKa = 10.6QNQLFGGAHH596 pKa = 5.79ATLPSNAPLGSWNWTAVYY614 pKa = 9.13STGSAANVKK623 pKa = 8.85ATGLFSVMAAATLDD637 pKa = 3.7TVTTDD642 pKa = 3.41IADD645 pKa = 4.05LVDD648 pKa = 5.54DD649 pKa = 5.02ISDD652 pKa = 3.45ISDD655 pKa = 3.53DD656 pKa = 3.61VATINTNINNLEE668 pKa = 4.07DD669 pKa = 5.57LIDD672 pKa = 4.09ALEE675 pKa = 4.23VTVDD679 pKa = 3.26IPEE682 pKa = 4.46LTTLTNDD689 pKa = 3.09VASLKK694 pKa = 10.54VSVSALDD701 pKa = 3.69AVVTAVAGDD710 pKa = 3.75VATVDD715 pKa = 3.76TKK717 pKa = 10.87IGTLTGTITGIEE729 pKa = 4.15GNIATIEE736 pKa = 4.17TDD738 pKa = 3.23VGTLQADD745 pKa = 3.48ISDD748 pKa = 3.82VEE750 pKa = 4.67ANVDD754 pKa = 3.73STPAWIAVVLALVAAVAAIFAVITIRR780 pKa = 11.84QKK782 pKa = 10.44IAGG785 pKa = 3.82
Molecular weight: 79.83 kDa
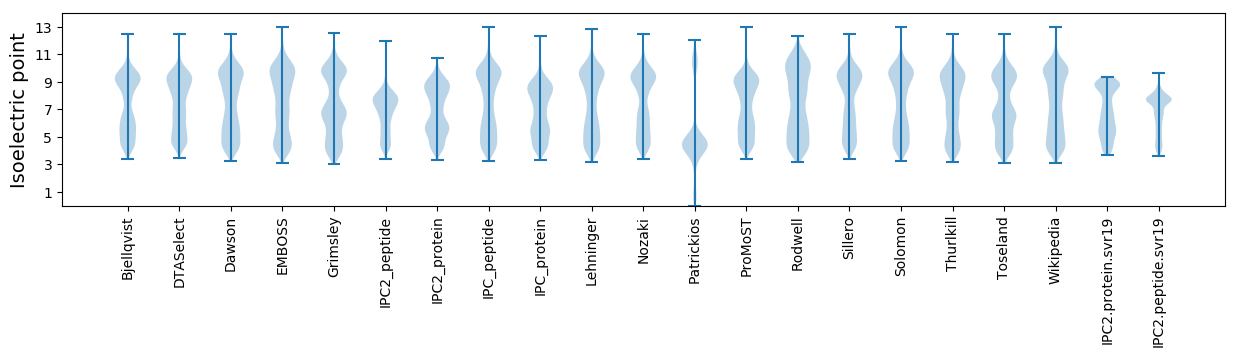
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U0RZ30|A0A2U0RZ30_9ARCH Aspartate aminotransferase family protein OS=Candidatus Bathyarchaeota archaeon OX=2026714 GN=CW691_07835 PE=3 SV=1
MM1 pKa = 7.81RR2 pKa = 11.84LAKK5 pKa = 10.19AGKK8 pKa = 8.64EE9 pKa = 3.98KK10 pKa = 10.84KK11 pKa = 9.46PVPTWVIAKK20 pKa = 8.97TGGQVRR26 pKa = 11.84SNPKK30 pKa = 8.73RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84WRR35 pKa = 11.84QRR37 pKa = 11.84KK38 pKa = 8.35IRR40 pKa = 11.84AA41 pKa = 3.38
MM1 pKa = 7.81RR2 pKa = 11.84LAKK5 pKa = 10.19AGKK8 pKa = 8.64EE9 pKa = 3.98KK10 pKa = 10.84KK11 pKa = 9.46PVPTWVIAKK20 pKa = 8.97TGGQVRR26 pKa = 11.84SNPKK30 pKa = 8.73RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84WRR35 pKa = 11.84QRR37 pKa = 11.84KK38 pKa = 8.35IRR40 pKa = 11.84AA41 pKa = 3.38
Molecular weight: 4.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
642103 |
25 |
1519 |
275.7 |
30.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.205 ± 0.047 | 1.285 ± 0.025 |
5.192 ± 0.046 | 6.588 ± 0.051 |
4.191 ± 0.039 | 6.954 ± 0.055 |
1.691 ± 0.023 | 7.18 ± 0.049 |
6.925 ± 0.074 | 9.303 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.359 ± 0.024 | 4.462 ± 0.048 |
4.03 ± 0.031 | 3.071 ± 0.029 |
4.093 ± 0.043 | 6.475 ± 0.049 |
6.387 ± 0.061 | 7.878 ± 0.044 |
1.268 ± 0.025 | 3.462 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
